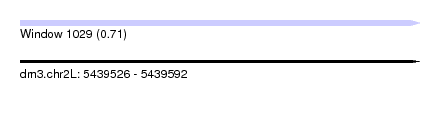
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,439,526 – 5,439,592 |
| Length | 66 |
| Max. P | 0.706895 |

| Location | 5,439,526 – 5,439,592 |
|---|---|
| Length | 66 |
| Sequences | 11 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 84.89 |
| Shannon entropy | 0.33431 |
| G+C content | 0.41067 |
| Mean single sequence MFE | -15.26 |
| Consensus MFE | -12.67 |
| Energy contribution | -11.75 |
| Covariance contribution | -0.93 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.706895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5439526 66 + 23011544 AAGUGGAAAGUGACAGGCGAGUGUCGCUUUUAUUGUUUACUUUCAAUCACUUUUCGUCACGAAUUC-- ..(((((((((((.(((((.....)))))...(((........))))))))))))).)........-- ( -15.30, z-score = -1.31, R) >droPer1.super_5 6336846 66 - 6813705 CAGUGGAAAGUGACGGUAGAGUGUCGCUUUUAUUGUUUACUUUCAAUCACUUUUCGUCACGAAUUC-- ..(((((((((((..(.((((((..((.......)).)))))))..)))))))))).)........-- ( -15.80, z-score = -1.66, R) >dp4.chr4_group1 4741161 66 + 5278887 CAGUGGAAAGUGACGGUAGAGUGUCGCUUUUAUUGUUUACUUUCAAUCACUUUUCGUCACGAAUUC-- ..(((((((((((..(.((((((..((.......)).)))))))..)))))))))).)........-- ( -15.80, z-score = -1.66, R) >droAna3.scaffold_12916 6466386 66 - 16180835 AAGUGGAAAGUGACGGGAUAGUGUCGCUUUUAUUGUUUACUUUCAAUCACUUUUCGUCACGAAUUC-- ..(((((((((((..(((.((((..((.......)).)))))))..)))))))))).)........-- ( -15.20, z-score = -1.58, R) >droEre2.scaffold_4929 5523552 66 + 26641161 AAGUGGAAAGUGACAGGCGAGUGUCGCUUUUAUUGUUUACUUUCAAUCACUUUUCGUCACGAAUUC-- ..(((((((((((.(((((.....)))))...(((........))))))))))))).)........-- ( -15.30, z-score = -1.31, R) >droYak2.chr2L 8566454 66 - 22324452 AAGUGGAAAGUGACAGGCGAGUGUCGCUUUUAUUGUUUACUUUCAAUCACUUUUCGUCACGAAUUC-- ..(((((((((((.(((((.....)))))...(((........))))))))))))).)........-- ( -15.30, z-score = -1.31, R) >droSec1.super_5 3513839 66 + 5866729 AAGUGGAAAGUGACAGGCGAGUGUCGCUUUUAUUGUUUACUUUCAAUCACUUUUCGUCACGAAUUC-- ..(((((((((((.(((((.....)))))...(((........))))))))))))).)........-- ( -15.30, z-score = -1.31, R) >droSim1.chr2L 5242295 66 + 22036055 AAGUGGAAAGUGACAGGCGAGUGUCGCUUUUAUUGUUUACUUUCAAUCACUUUUCGUCACGAAUUC-- ..(((((((((((.(((((.....)))))...(((........))))))))))))).)........-- ( -15.30, z-score = -1.31, R) >droVir3.scaffold_12963 375190 67 - 20206255 AGGUGGAAAGUGGCG-UCGAGUGUCGCUUUUAUUGUUUACUUUCAAUCACUUUUCGUCACGAAGUAAC .(..(((((((((..-..(((((..((.......)).)))))....)))))))))..).......... ( -15.10, z-score = -0.97, R) >droMoj3.scaffold_6500 23362835 67 + 32352404 AGGUGGAAAGUGGCG-UCGAGUGUCGCUUUUAUUGUUUACUUUCAAUCACAUUUCGUCACGAAGCAAC ..(((((((((((((-.....))))))))))((((........))))))).(((((...))))).... ( -13.40, z-score = -0.15, R) >anoGam1.chrX 2507648 66 + 22145176 ACCCAGACAGAUAGAGGCGCGUGUGACAGUAUUUGCGUGUUGGCGGACAGUUUUCGUUCGCAGUUC-- .....(((.....((.(((((.(((....))).))))).)).((((((.......)))))).))).-- ( -16.10, z-score = 0.19, R) >consensus AAGUGGAAAGUGACAGGCGAGUGUCGCUUUUAUUGUUUACUUUCAAUCACUUUUCGUCACGAAUUC__ ..(((((((((((((......))))))))))((((........))))..........)))........ (-12.67 = -11.75 + -0.93)

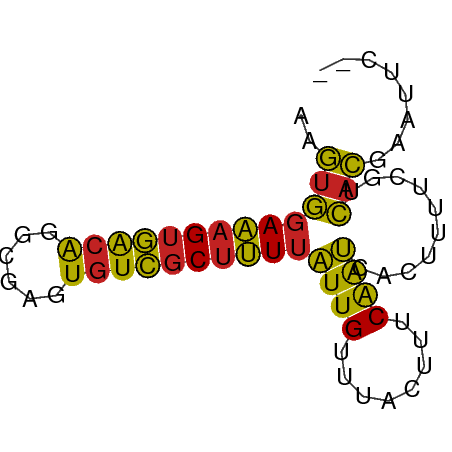
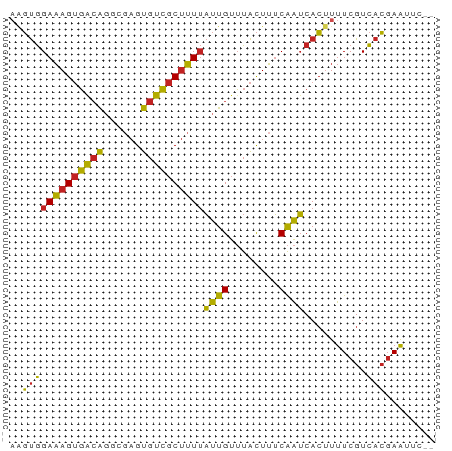
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:00 2011