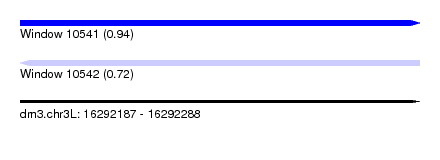
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,292,187 – 16,292,288 |
| Length | 101 |
| Max. P | 0.941713 |

| Location | 16,292,187 – 16,292,288 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 62.91 |
| Shannon entropy | 0.51171 |
| G+C content | 0.50258 |
| Mean single sequence MFE | -32.11 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
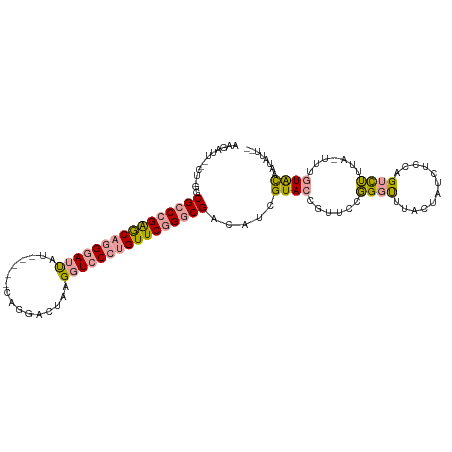
>dm3.chr3L 16292187 101 + 24543557 -------------CGGCCGAACAGGGACCGUAUGGUCAGGACUAAGAUCCCUGUUCGGGCGCCAUCGUACCGUUCCUGUUCUACUCUCUGCAGUUACUC-UCCAUAUAACCUGCG -------------((.(((((((((((.(...((((....)))).).))))))))))).))..........((..((((..........))))..))..-............... ( -31.50, z-score = -1.52, R) >droEre2.scaffold_4784 18534847 107 + 25762168 AAGAUUAACU-GGCGCCCGGGCAGGGAUUAU-----CAGGACUAAGGUCCCUGUUCGGGCGUCAUCGUACUGUUCCGGGCUUUCUGUCUCCAGUCUUCAGUUUGUGCAAUAUU-- .........(-((((((((((((((((((..-----.........)))))))))))))))))))..((((......(((((..........))))).......))))......-- ( -40.82, z-score = -3.24, R) >droYak2.chr3L 2293900 107 - 24197627 AAGAUUGUCUUGGCGCCCGAGCAGGAAUUAU-----CAGGAAUAGGGUCCA-GUUCAGGCGAAUUCGUACGGUUUCGGGUUUCCUAUCUCCAGUCUUUAAUUUGUACAAUAUU-- (((((((...(((((((.((((.(((.((((-----.....))))..))).-)))).))))((((((........))))))..)))....)))))))................-- ( -24.00, z-score = 0.20, R) >consensus AAGAUU__CU_GGCGCCCGAGCAGGGAUUAU_____CAGGACUAAGGUCCCUGUUCGGGCGACAUCGUACCGUUCCGGGUUUACUAUCUCCAGUCUUUA_UUUGUACAAUAUU__ .............((((((((((((((((................)))))))))))))))).....((((......((((............)))).......))))........ (-19.56 = -20.24 + 0.68)

| Location | 16,292,187 – 16,292,288 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 62.91 |
| Shannon entropy | 0.51171 |
| G+C content | 0.50258 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -15.63 |
| Energy contribution | -16.75 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.715388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
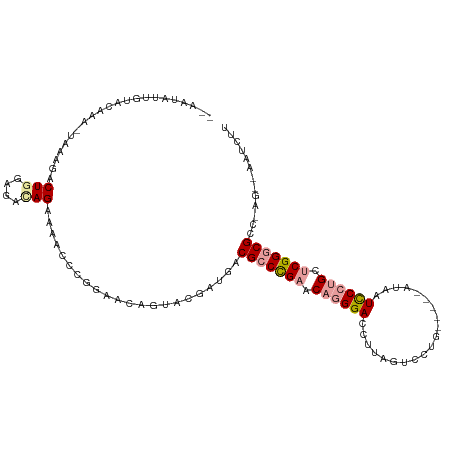
>dm3.chr3L 16292187 101 - 24543557 CGCAGGUUAUAUGGA-GAGUAACUGCAGAGAGUAGAACAGGAACGGUACGAUGGCGCCCGAACAGGGAUCUUAGUCCUGACCAUACGGUCCCUGUUCGGCCG------------- .((((.((((.....-..))))))))...(.((.(((((((...((.((...((..(((.....)))..))..)))).((((....))))))))))).))).------------- ( -32.20, z-score = -0.87, R) >droEre2.scaffold_4784 18534847 107 - 25762168 --AAUAUUGCACAAACUGAAGACUGGAGACAGAAAGCCCGGAACAGUACGAUGACGCCCGAACAGGGACCUUAGUCCUG-----AUAAUCCCUGCCCGGGCGCC-AGUUAAUCUU --...........(((((....(((....)))...((((((.......((........))..((((((..(((....))-----)...)))))).))))))..)-))))...... ( -32.80, z-score = -2.82, R) >droYak2.chr3L 2293900 107 + 24197627 --AAUAUUGUACAAAUUAAAGACUGGAGAUAGGAAACCCGAAACCGUACGAAUUCGCCUGAAC-UGGACCCUAUUCCUG-----AUAAUUCCUGCUCGGGCGCCAAGACAAUCUU --....((((((...................((....))(....)))))))...(((((((.(-.(((...((((...)-----)))..))).).)))))))............. ( -18.90, z-score = 0.13, R) >consensus __AAUAUUGUACAAA_UAAAGACUGGAGACAGAAAACCCGGAACAGUACGAUGACGCCCGAACAGGGACCUUAGUCCUG_____AUAAUCCCUGCUCGGGCGCC_AG__AAUCUU ......................(((....)))......................(((((((.((((((....................)))))).)))))))............. (-15.63 = -16.75 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:31:03 2011