| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,242,138 – 16,242,229 |
| Length | 91 |
| Max. P | 0.875435 |

| Location | 16,242,138 – 16,242,229 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 70.90 |
| Shannon entropy | 0.58045 |
| G+C content | 0.42886 |
| Mean single sequence MFE | -21.71 |
| Consensus MFE | -9.62 |
| Energy contribution | -8.78 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16242138 91 + 24543557 -UCCCCAUCAAGAUAUCUACACAGCGUCUUGACAACCGAAAACGAGUGGUAACGGAGCAACAGAGAGAUAUCUUGAUGG-CAAUUCAAUCGUU -...((((((((((((((.....((.(((..((.((((....)).)).))...))))).......))))))))))))))-............. ( -26.10, z-score = -3.19, R) >droSec1.super_0 8336233 91 + 21120651 -UCCCCAUCAAGAUAUCGACACAGCGUCUUGACAACCGAAAACGAGUGGUAACGGAGCAACAGAGAGAUAUCUUGAUGG-CAAUUCAAUCGUU -...((((((((((((((((.....))).......(((...((.....))..)))...........)))))))))))))-............. ( -24.80, z-score = -2.49, R) >droSim1.chr3L 15577189 91 + 22553184 -UCCCCAUCAAGAUAUCCACACAGCGUCUUGACAACCGAAAACGAGUGGUAACGGAGCAACAGAGAGAUAUCUUGAUGG-CAAUUCAAUCGUU -...((((((((((((((.(...((.(((..((.((((....)).)).))...)))))....).).)))))))))))))-............. ( -23.70, z-score = -2.41, R) >droYak2.chr3L 2248562 91 - 24197627 -UCCCCAUCAAGAUAUCUACACAGCGUCUUGACAACCGAAAACGAGUGGUAACGGAGCAACAGAGGGAUAUCUUGAUGG-CAAUUCAAUCGUU -...((((((((((((((.....((.(((..((.((((....)).)).))...))))).......))))))))))))))-............. ( -26.10, z-score = -2.63, R) >droEre2.scaffold_4784 18484903 91 + 25762168 -UCCCCAUCAAGAUAUCUGCACAGCGUCUUGACAACCGAAAACGAGUGGUAACGAAGCAACAGAGGUAUAUCUUGAUGG-CAAUUCAAUCGUU -...((((((((((((((.....((.((...((.((((....)).)).))...)).))......)).))))))))))))-............. ( -22.40, z-score = -1.67, R) >droAna3.scaffold_13337 12905905 89 - 23293914 UCCCGCAUCAAGAUUUCCAGCCAUUUCGCUGCCU-UGACAACCGAAUAGUAACGGGGCGACACA--CAAAUCCUGAUGG-CAAUUCAAUCGGU ..((((((((.(((((..(((......)))((((-((...((......))..))))))......--.))))).))))).-.........))). ( -17.60, z-score = 0.45, R) >droVir3.scaffold_13049 1215048 85 + 25233164 -UUUCCAUAAACAUUUUAUGGCGUUUGCCUGGCAACGAUUUCCAUCGGGCAAUGUAUGAA-------AAAUAUUGAUGGUUCUUUCAAUCGCU -...((((.....(((((((.((((.(((((....)(((....)))))))))))))))))-------).......)))).............. ( -15.80, z-score = 0.21, R) >droGri2.scaffold_15110 2346188 82 + 24565398 -UUUCCAUAAACAUUUUAUGGCGUUUGCUUGGCAACGAUAUCCAUCAG-UAAUACAUGAA-------AAAU--UGAUGGGUUUUUCAAUCGUU -...((((((.....)))))).(((.....)))(((((((((((((((-(..........-------..))--))))))))......)))))) ( -17.20, z-score = -1.48, R) >consensus _UCCCCAUCAAGAUAUCUACACAGCGUCUUGACAACCGAAAACGAGUGGUAACGGAGCAACAGAG_GAUAUCUUGAUGG_CAAUUCAAUCGUU ....((((((((((((.........((.(((....((........).)....))).)).........)))))))))))).............. ( -9.62 = -8.78 + -0.84)

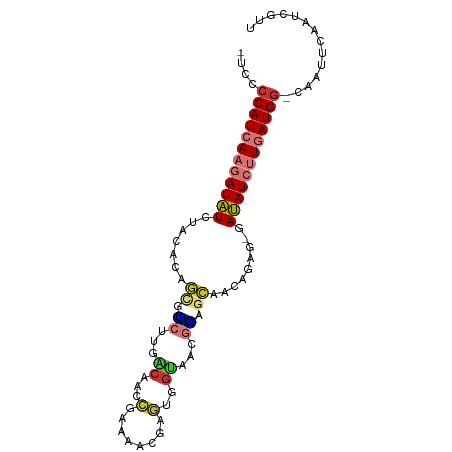
| Location | 16,242,138 – 16,242,229 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 70.90 |
| Shannon entropy | 0.58045 |
| G+C content | 0.42886 |
| Mean single sequence MFE | -23.23 |
| Consensus MFE | -10.91 |
| Energy contribution | -10.79 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.699372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16242138 91 - 24543557 AACGAUUGAAUUG-CCAUCAAGAUAUCUCUCUGUUGCUCCGUUACCACUCGUUUUCGGUUGUCAAGACGCUGUGUAGAUAUCUUGAUGGGGA- ..........((.-((((((((((((((......(((..((((((.((.((....)))).))...))))..))).)))))))))))))).))- ( -25.80, z-score = -1.70, R) >droSec1.super_0 8336233 91 - 21120651 AACGAUUGAAUUG-CCAUCAAGAUAUCUCUCUGUUGCUCCGUUACCACUCGUUUUCGGUUGUCAAGACGCUGUGUCGAUAUCUUGAUGGGGA- ..........((.-(((((((((((((......((((.(((..((.....))...)))..).)))(((.....)))))))))))))))).))- ( -26.80, z-score = -1.93, R) >droSim1.chr3L 15577189 91 - 22553184 AACGAUUGAAUUG-CCAUCAAGAUAUCUCUCUGUUGCUCCGUUACCACUCGUUUUCGGUUGUCAAGACGCUGUGUGGAUAUCUUGAUGGGGA- ..........((.-(((((((((((((..(((...((.(((..((.....))...)))..))..)))(((...)))))))))))))))).))- ( -25.90, z-score = -1.43, R) >droYak2.chr3L 2248562 91 + 24197627 AACGAUUGAAUUG-CCAUCAAGAUAUCCCUCUGUUGCUCCGUUACCACUCGUUUUCGGUUGUCAAGACGCUGUGUAGAUAUCUUGAUGGGGA- ..........((.-(((((((((((((.....((.((...)).))(((.((((((........))))))..)))..))))))))))))).))- ( -24.50, z-score = -1.26, R) >droEre2.scaffold_4784 18484903 91 - 25762168 AACGAUUGAAUUG-CCAUCAAGAUAUACCUCUGUUGCUUCGUUACCACUCGUUUUCGGUUGUCAAGACGCUGUGCAGAUAUCUUGAUGGGGA- ..........((.-(((((((((((....(((((.((.((...((.((.((....)))).))...)).))...)))))))))))))))).))- ( -27.60, z-score = -2.29, R) >droAna3.scaffold_13337 12905905 89 + 23293914 ACCGAUUGAAUUG-CCAUCAGGAUUUG--UGUGUCGCCCCGUUACUAUUCGGUUGUCA-AGGCAGCGAAAUGGCUGGAAAUCUUGAUGCGGGA .(((.........-.((((((((((((--((...))).(((...(((((..(((((..-..)))))..))))).))))))))))))))))).. ( -23.60, z-score = 0.43, R) >droVir3.scaffold_13049 1215048 85 - 25233164 AGCGAUUGAAAGAACCAUCAAUAUUU-------UUCAUACAUUGCCCGAUGGAAAUCGUUGCCAGGCAAACGCCAUAAAAUGUUUAUGGAAA- ..............((((.(((((((-------(.......(((((((((....))))......))))).......)))))))).))))...- ( -18.44, z-score = -1.18, R) >droGri2.scaffold_15110 2346188 82 - 24565398 AACGAUUGAAAAACCCAUCA--AUUU-------UUCAUGUAUUA-CUGAUGGAUAUCGUUGCCAAGCAAACGCCAUAAAAUGUUUAUGGAAA- ...((((((........)))--)))(-------((((((.((..-...((((......((((...))))...)))).....)).))))))).- ( -13.20, z-score = -0.40, R) >consensus AACGAUUGAAUUG_CCAUCAAGAUAUC_CUCUGUUGCUCCGUUACCACUCGUUUUCGGUUGUCAAGACGCUGUGUAGAUAUCUUGAUGGGGA_ ..............((((((((((((...................................................)))))))))))).... (-10.91 = -10.79 + -0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:59 2011