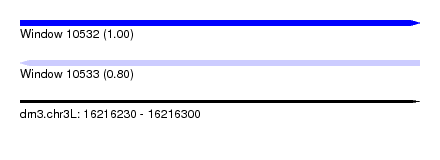
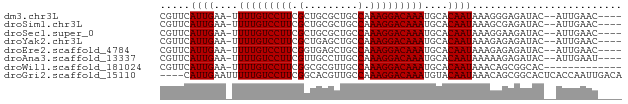
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,216,230 – 16,216,300 |
| Length | 70 |
| Max. P | 0.999026 |

| Location | 16,216,230 – 16,216,300 |
|---|---|
| Length | 70 |
| Sequences | 8 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 84.27 |
| Shannon entropy | 0.30907 |
| G+C content | 0.41618 |
| Mean single sequence MFE | -22.53 |
| Consensus MFE | -17.71 |
| Energy contribution | -17.96 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.72 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
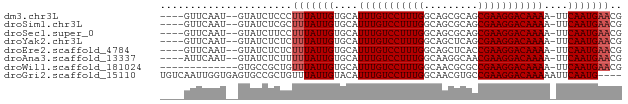
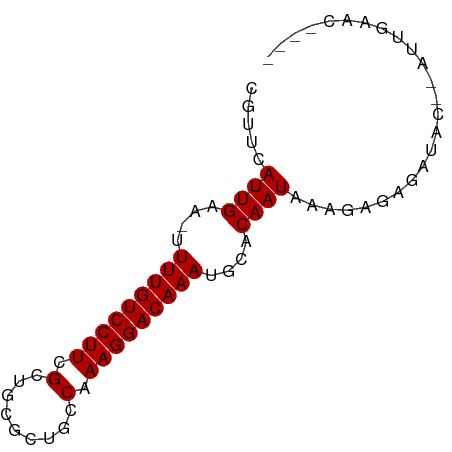
>dm3.chr3L 16216230 70 + 24543557 ----GUUCAAU--GUAUCUCCCUUUAUUGUGCAUUUGUCCUUUGGCAGCGCAGCGAAGGACAAAA-UUCAAUGAACG ----(((((.(--(......((((..((((((...((((....))))))))))..))))......-..)).))))). ( -20.74, z-score = -2.99, R) >droSim1.chr3L 15549791 70 + 22553184 ----GUUCAAU--GUAUCUCGCUUUAUUGUGCAUUUGUCCUUUGGCAGCGCAGCGAAGGACAAAA-UUCAAUGAACG ----(((((.(--(......((........)).(((((((((((.(......)))))))))))).-..)).))))). ( -21.40, z-score = -2.67, R) >droSec1.super_0 8310559 70 + 21120651 ----GUUCAAU--GUAUCUUCCUUUAUUGUGCAUUUGUCCUUUGGCAGCGCAGCGAAGGACAAAA-UUCAAUGAACG ----(((((.(--(.....(((((..((((((...((((....))))))))))..))))).....-..)).))))). ( -22.60, z-score = -3.70, R) >droYak2.chr3L 2222700 70 - 24197627 ----GUUCAAU--GUAUCUCUCUUUAUUGUGCAUUUGUCCUUUGGCAGCUCAGCGAAGGACAAAA-UUCAAUGAACG ----(((((.(--((((...........)))))(((((((((((.(......)))))))))))).-.....))))). ( -19.90, z-score = -3.21, R) >droEre2.scaffold_4784 18459211 70 + 25762168 ----GUUCAAU--GUAUCUCUCUUUAUUGUGCAUUUGUCCUUUGGCAGCUCACCGAAGGACAAAA-UUCAAUGAACG ----(((((.(--((((...........)))))((((((((((((.......)))))))))))).-.....))))). ( -22.70, z-score = -4.87, R) >droAna3.scaffold_13337 12880258 70 - 23293914 ----AUUCAAU--GUAUCUCUUUUUAUUGUGCAUUUGUCCUUUGGCAAGGCAACGAAGGACAAAA-UUCAAUGAACG ----.......--.........(((((((....(((((((((((.........))))))))))).-..))))))).. ( -18.30, z-score = -3.03, R) >droWil1.scaffold_181024 69363 63 - 1751709 -------------GUGCCGCUGUUUAUUGUGCAUUUGUCCUUUGGCAACGCGCCGAAGGACAAAA-UUCAAUGAACG -------------........((((((((....(((((((((((((.....))))))))))))).-..)))))))). ( -28.60, z-score = -5.47, R) >droGri2.scaffold_15110 2320273 73 + 24565398 UGUCAAUUGGUGAGUGCCGCUGUUUAUUGUACAUUUGUCCUUUGGCAACGUGCCGAAGGACAAAAAUUCAAUG---- (((((((.((((.....))))....)))).)))(((((((((((((.....))))))))))))).........---- ( -26.00, z-score = -3.84, R) >consensus ____GUUCAAU__GUAUCUCUCUUUAUUGUGCAUUUGUCCUUUGGCAGCGCAGCGAAGGACAAAA_UUCAAUGAACG ......................(((((((....(((((((((((.........)))))))))))....))))))).. (-17.71 = -17.96 + 0.25)
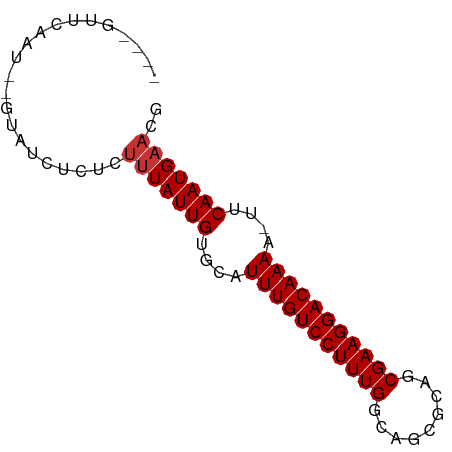
| Location | 16,216,230 – 16,216,300 |
|---|---|
| Length | 70 |
| Sequences | 8 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 84.27 |
| Shannon entropy | 0.30907 |
| G+C content | 0.41618 |
| Mean single sequence MFE | -17.29 |
| Consensus MFE | -11.16 |
| Energy contribution | -11.16 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.802656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16216230 70 - 24543557 CGUUCAUUGAA-UUUUGUCCUUCGCUGCGCUGCCAAAGGACAAAUGCACAAUAAAGGGAGAUAC--AUUGAAC---- .(((((.((..-.(((((((((.((......))..)))))))))....(......).......)--).)))))---- ( -15.80, z-score = -1.10, R) >droSim1.chr3L 15549791 70 - 22553184 CGUUCAUUGAA-UUUUGUCCUUCGCUGCGCUGCCAAAGGACAAAUGCACAAUAAAGCGAGAUAC--AUUGAAC---- .(((((.((..-.(((((((((.((......))..)))))))))(((........))).....)--).)))))---- ( -19.20, z-score = -2.49, R) >droSec1.super_0 8310559 70 - 21120651 CGUUCAUUGAA-UUUUGUCCUUCGCUGCGCUGCCAAAGGACAAAUGCACAAUAAAGGAAGAUAC--AUUGAAC---- .(((((.((..-.(((((((((.((......))..)))))))))....(......).......)--).)))))---- ( -15.80, z-score = -1.43, R) >droYak2.chr3L 2222700 70 + 24197627 CGUUCAUUGAA-UUUUGUCCUUCGCUGAGCUGCCAAAGGACAAAUGCACAAUAAAGAGAGAUAC--AUUGAAC---- .(((((.((..-.(((((((((.((......))..)))))))))....(......).......)--).)))))---- ( -15.60, z-score = -1.70, R) >droEre2.scaffold_4784 18459211 70 - 25762168 CGUUCAUUGAA-UUUUGUCCUUCGGUGAGCUGCCAAAGGACAAAUGCACAAUAAAGAGAGAUAC--AUUGAAC---- .(((((.((..-.(((((((((.(((.....))).)))))))))....(......).......)--).)))))---- ( -18.40, z-score = -2.61, R) >droAna3.scaffold_13337 12880258 70 + 23293914 CGUUCAUUGAA-UUUUGUCCUUCGUUGCCUUGCCAAAGGACAAAUGCACAAUAAAAAGAGAUAC--AUUGAAU---- .(((((.((..-.(((((((((.((......))..)))))))))....(........).....)--).)))))---- ( -12.00, z-score = -0.82, R) >droWil1.scaffold_181024 69363 63 + 1751709 CGUUCAUUGAA-UUUUGUCCUUCGGCGCGUUGCCAAAGGACAAAUGCACAAUAAACAGCGGCAC------------- .(((.((((..-.(((((((((.(((.....))).)))))))))....)))).)))........------------- ( -20.70, z-score = -2.86, R) >droGri2.scaffold_15110 2320273 73 - 24565398 ----CAUUGAAUUUUUGUCCUUCGGCACGUUGCCAAAGGACAAAUGUACAAUAAACAGCGGCACUCACCAAUUGACA ----.((((.((.(((((((((.(((.....))).))))))))).)).)))).......((......))........ ( -20.80, z-score = -3.09, R) >consensus CGUUCAUUGAA_UUUUGUCCUUCGCUGCGCUGCCAAAGGACAAAUGCACAAUAAAGAGAGAUAC__AUUGAAC____ .....((((....(((((((((.(.........).)))))))))....))))......................... (-11.16 = -11.16 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:56 2011