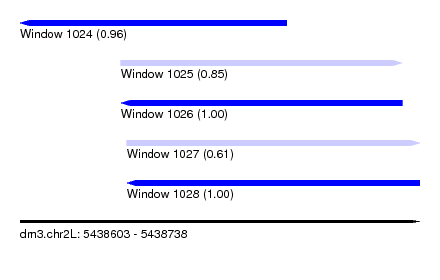
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,438,603 – 5,438,738 |
| Length | 135 |
| Max. P | 0.999427 |

| Location | 5,438,603 – 5,438,693 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.92 |
| Shannon entropy | 0.36665 |
| G+C content | 0.42644 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -16.97 |
| Energy contribution | -17.26 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5438603 90 - 23011544 AAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUC-------UCUGUCUCCGAUUUUUUGUGUAUUUUCGUG-AC--UCUCAACUG ..(((((...))))).....(((((((((((....)))))))))))...-------...(((..(((..............))).)-))--......... ( -21.14, z-score = -1.76, R) >droSim1.chr2L 5241368 92 - 22036055 AAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUC-------UCUGUCUCCGAUUUUUUGUGUUUUUUCCUG-ACUGUGUUAACUG .(((((...(((.(.((...(((((((((((....)))))))))))...-------..)).).))).........(((.......)-)).)))))..... ( -21.00, z-score = -1.66, R) >droSec1.super_5 3512911 93 - 5866729 AAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUC-------UCUGUCUCCGAUUUUUUGUGUUUUUUCCUACUCUGUGUUAACUG .(((((..((((........(((((((((((....)))))))))))...-------........(((....))).........))))...)))))..... ( -21.00, z-score = -2.18, R) >droYak2.chr2L 8565564 89 + 22324452 AAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUC-------UCUGUCUCCGAUUUUUUGUGUA-UUUCGUG-AC--UGUUAACUG ..(((((...))))).....(((((((((((....)))))))))))...-------...(((..(((...........-..))).)-))--......... ( -21.22, z-score = -1.60, R) >droEre2.scaffold_4929 5522669 76 - 26641161 AAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUC-------UCUGUCUCCGAUUUGUUGUGUAUUUUG----------------- ..((((...(((.(.((...(((((((((((....)))))))))))...-------..)).).))).......))))......----------------- ( -19.70, z-score = -2.10, R) >droAna3.scaffold_12916 6465557 90 + 16180835 AAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUC-------UCUGUCUCCGGUUUGUUGAUUUUUUUUUU---UUUUGGACACUC ..........((((((......(((((((((....)))))))))(((..-------.(((....)))...)))............---.....)))))). ( -22.50, z-score = -1.99, R) >dp4.chr4_group1 4739949 94 - 5278887 AAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCAUCAGAAUCUGUCUCUGGAUAUUGGAAGUUUCUCUCUCUCUCUCU------ ...((((...))))(((((((((((((((((....)))))))))).....(((((.......))))))))))))....................------ ( -23.40, z-score = -1.63, R) >consensus AAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUC_______UCUGUCUCCGAUUUUUUGUGUUUUUUCCUG_ACU_UGUUAACUG ..(((((...))))).....(((((((((((....)))))))))))...................................................... (-16.97 = -17.26 + 0.29)

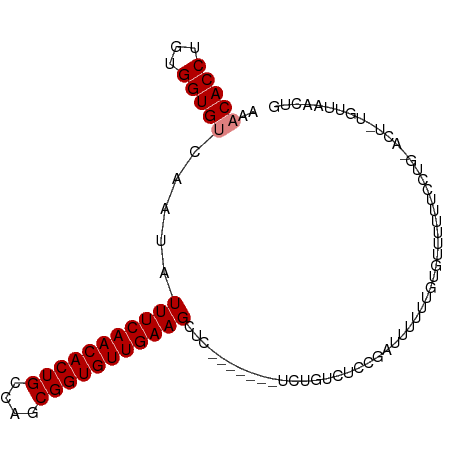
| Location | 5,438,637 – 5,438,732 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.01 |
| Shannon entropy | 0.55044 |
| G+C content | 0.43545 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -12.44 |
| Energy contribution | -12.54 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5438637 95 + 23011544 ---------------AGACAGAGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGUUUUUACUUGGAAGUCAAGAGGUUGAUUUGCCUUAGA-CUUAG ---------------.....(((...((((((((.(....).)))))))).((..((((((...))))))..)))))..(((((..(((((......))))).))-))).. ( -26.30, z-score = -1.53, R) >droSim1.chr2L 5241404 90 + 22036055 ---------------AGACAGAGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGUUUUUACUUGGAA-----GAGGUUGAUUUGCCUUAGA-CUUAG ---------------.....(((.((((((((((.(....).))))))))..((..((..(.((((((....))))))...-----)..))..))..)))))...-..... ( -21.90, z-score = -0.78, R) >droSec1.super_5 3512948 95 + 5866729 ---------------AGACAGAGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGUUUUUACUUGGAAGUCAAGAGGUUGAUUUGCCUUAGA-CUUAG ---------------.....(((...((((((((.(....).)))))))).((..((((((...))))))..)))))..(((((..(((((......))))).))-))).. ( -26.30, z-score = -1.53, R) >droYak2.chr2L 8565597 95 - 22324452 ---------------AGACAGAGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGUUUUUACUGGGAAGUCAAGAGGUUGAUUUGCCUUAGA-CUUAG ---------------...(((.....((((((((.(....).)))))))).((..((((((...))))))..)))))..(((((..(((((......))))).))-))).. ( -28.40, z-score = -2.00, R) >droEre2.scaffold_4929 5522689 95 + 26641161 ---------------AGACAGAGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGUUUUUACUGGGAAGUCGAGAGGUUGAUUUGCCUUAGA-CUUAG ---------------...(((.....((((((((.(....).)))))))).((..((((((...))))))..)))))..(((((..(((((......))))).))-))).. ( -29.80, z-score = -2.46, R) >droAna3.scaffold_12916 6465591 96 - 16180835 ---------------AGACAGAGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGAUUUCGGAGGAAAGUCGAGAGGUCAGUUGGCUUAAAAACCCAA ---------------.......((((((((((((.(....).))))))))............(((.(((((((.((......))..))))))).))))))).......... ( -24.20, z-score = -0.49, R) >dp4.chr4_group1 4739980 98 + 5278887 ------AGACAGAUU--CUGAUGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGAUUUUAUAUGGAAGUC----AGUUAGAGCCCCCCAGACUUGG- ------...(((..(--(((..(.(((..(((....(((...((((..(....)..))))..))).(((((((.....))))))----))))..))).)..)))).))).- ( -24.70, z-score = -0.98, R) >droPer1.super_5 6335687 98 - 6813705 ------AGACAGAUU--CUGAUGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGAUUUUAUAUGGAAGUC----AGUUAGAGCCCCCCAGACUAGG- ------........(--(((..(.(((..(((....(((...((((..(....)..))))..))).(((((((.....))))))----))))..))).)..)))).....- ( -24.10, z-score = -0.95, R) >droWil1.scaffold_180708 582982 95 + 12563649 -----ACAGUGUCUCAACUGAUGAACUUCAACACUGCUGACAGUGUUGAAAUAUUGACACCACAGGUGAUUUCAUAUGGAAGUC----AUUUCUUUGUUGUUUU------- -----.((((......))))..((((((((((((((....)))))))))).....((((....((((((((((.....))))))----))))...)))))))).------- ( -27.70, z-score = -2.05, R) >droVir3.scaffold_12963 373298 103 - 20206255 GGGCAGAAACAGCUCAGCUGAUGAGCAUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGAUUUUAUAUGGAAGUC----AAUAAUUUGUUUUUGUUUG---- ((((((((((((((((.....))))).(((((((.(....).)))))))..(((((((.(((...(((....))).)))..)))----))))...))))))))))).---- ( -33.40, z-score = -3.14, R) >consensus _______________AGACAGAGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGAUUUUACAUGGAAGUC__GAGGUUGAUUUGCCUUAGA_CUUAG ..........................((((((((.(....).)))))))).......((((...))))........................................... (-12.44 = -12.54 + 0.10)

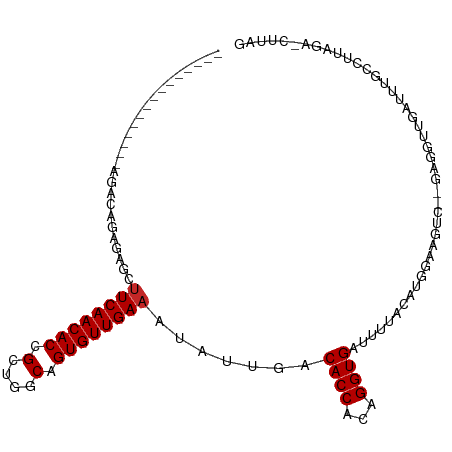
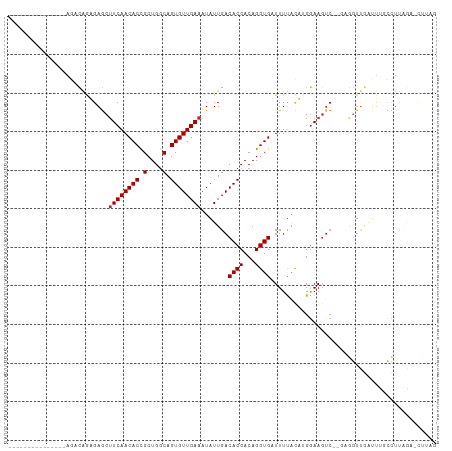
| Location | 5,438,637 – 5,438,732 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.01 |
| Shannon entropy | 0.55044 |
| G+C content | 0.43545 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -16.70 |
| Energy contribution | -16.71 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5438637 95 - 23011544 CUAAG-UCUAAGGCAAAUCAACCUCUUGACUUCCAAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCUCUGUCU--------------- .....-....(((((..........(((((..(((.((......))...))).)))))..(((((((((((....))))))))))).....)))))--------------- ( -24.60, z-score = -2.63, R) >droSim1.chr2L 5241404 90 - 22036055 CUAAG-UCUAAGGCAAAUCAACCUC-----UUCCAAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCUCUGUCU--------------- .....-....(((((..........-----............(((((...))))).....(((((((((((....))))))))))).....)))))--------------- ( -21.80, z-score = -2.47, R) >droSec1.super_5 3512948 95 - 5866729 CUAAG-UCUAAGGCAAAUCAACCUCUUGACUUCCAAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCUCUGUCU--------------- .....-....(((((..........(((((..(((.((......))...))).)))))..(((((((((((....))))))))))).....)))))--------------- ( -24.60, z-score = -2.63, R) >droYak2.chr2L 8565597 95 + 22324452 CUAAG-UCUAAGGCAAAUCAACCUCUUGACUUCCCAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCUCUGUCU--------------- .....-....(((((..........(((((..(((((.........))).)).)))))..(((((((((((....))))))))))).....)))))--------------- ( -26.00, z-score = -3.06, R) >droEre2.scaffold_4929 5522689 95 - 26641161 CUAAG-UCUAAGGCAAAUCAACCUCUCGACUUCCCAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCUCUGUCU--------------- .....-....(((((............(((..(((((.........))).)).)))....(((((((((((....))))))))))).....)))))--------------- ( -24.80, z-score = -2.91, R) >droAna3.scaffold_12916 6465591 96 + 16180835 UUGGGUUUUUAAGCCAACUGACCUCUCGACUUUCCUCCGAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCUCUGUCU--------------- ...((((....))))............(((........((.((((....)))).))....(((((((((((....)))))))))))......))).--------------- ( -23.10, z-score = -1.10, R) >dp4.chr4_group1 4739980 98 - 5278887 -CCAAGUCUGGGGGGCUCUAACU----GACUUCCAUAUAAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCAUCAG--AAUCUGUCU------ -.....((((..((((......(----(((..(((((..........))))).))))....((((((((((....))))))))))))))..)))--)........------ ( -32.60, z-score = -3.25, R) >droPer1.super_5 6335687 98 + 6813705 -CCUAGUCUGGGGGGCUCUAACU----GACUUCCAUAUAAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCAUCAG--AAUCUGUCU------ -.....((((..((((......(----(((..(((((..........))))).))))....((((((((((....))))))))))))))..)))--)........------ ( -32.60, z-score = -3.25, R) >droWil1.scaffold_180708 582982 95 - 12563649 -------AAAACAACAAAGAAAU----GACUUCCAUAUGAAAUCACCUGUGGUGUCAAUAUUUCAACACUGUCAGCAGUGUUGAAGUUCAUCAGUUGAGACACUGU----- -------....((((.......(----(((..(((((((....))..))))).))))..((((((((((((....))))))))))))......)))).........----- ( -28.20, z-score = -2.78, R) >droVir3.scaffold_12963 373298 103 + 20206255 ----CAAACAAAAACAAAUUAUU----GACUUCCAUAUAAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAUGCUCAUCAGCUGAGCUGUUUCUGCCC ----....((.(((((...((((----(((..(((((..........))))).)))))))..(((((((((....))))))))).(((((.....)))))))))).))... ( -31.90, z-score = -4.40, R) >consensus CUAAG_UCUAAGGCAAAUCAACCUC__GACUUCCAAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCUCUGUCU_______________ ...........................................((((...))))......(((((((((((....)))))))))))......................... (-16.70 = -16.71 + 0.01)
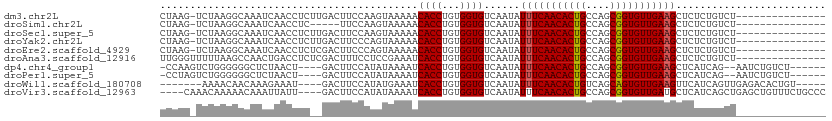
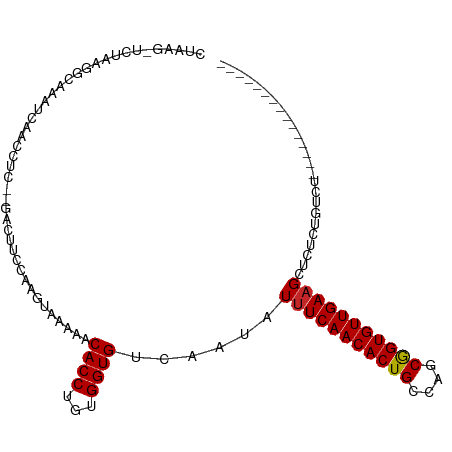
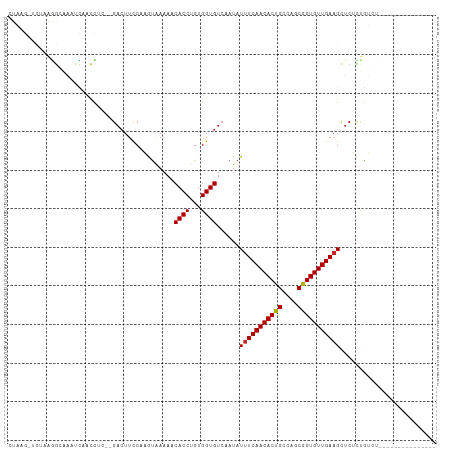
| Location | 5,438,639 – 5,438,738 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.42 |
| Shannon entropy | 0.57857 |
| G+C content | 0.43803 |
| Mean single sequence MFE | -26.65 |
| Consensus MFE | -12.29 |
| Energy contribution | -12.49 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.611531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5438639 99 + 23011544 ---------------ACAGAGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGUUUUUACUUGGAAGUCAAGAGGUUGAUUUGCCUUAGACUUAGGAACCU------ ---------------.........((((((((.(....).)))))))).........((.((((((....)))))).(((((..(((((......))))).))))).)).....------ ( -26.70, z-score = -1.15, R) >droSim1.chr2L 5241406 94 + 22036055 ---------------ACAGAGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGUUUUUACUUGG-----AAGAGGUUGAUUUGCCUUAGACUUAGGAACCU------ ---------------.........((((((((.(....).)))))))).........((.((((((....)))))).-----..(((((......))))).......)).....------ ( -22.20, z-score = -0.34, R) >droSec1.super_5 3512950 99 + 5866729 ---------------ACAGAGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGUUUUUACUUGGAAGUCAAGAGGUUGAUUUGCCUUAGACUUAGGAACCU------ ---------------.........((((((((.(....).)))))))).........((.((((((....)))))).(((((..(((((......))))).))))).)).....------ ( -26.70, z-score = -1.15, R) >droYak2.chr2L 8565599 99 - 22324452 ---------------ACAGAGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGUUUUUACUGGGAAGUCAAGAGGUUGAUUUGCCUUAGACUUAGGAACAU------ ---------------.(((.....((((((((.(....).)))))))).((..((((((...))))))..)))))..(((((..(((((......))))).)))))........------ ( -28.40, z-score = -1.78, R) >droEre2.scaffold_4929 5522691 99 + 26641161 ---------------ACAGAGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGUUUUUACUGGGAAGUCGAGAGGUUGAUUUGCCUUAGACUUAGGAACCU------ ---------------.(((.....((((((((.(....).)))))))).((..((((((...))))))..)))))..(((((..(((((......))))).)))))........------ ( -29.80, z-score = -1.89, R) >droAna3.scaffold_12916 6465593 105 - 16180835 ---------------ACAGAGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGAUUUCGGAGGAAAGUCGAGAGGUCAGUUGGCUUAAAAACCCAAGUCUUUAACCU ---------------.....((((((((((((.(....).))))))))............(((.(((((((.((......))..))))))).)))))))..................... ( -24.20, z-score = 0.31, R) >dp4.chr4_group1 4739982 102 + 5278887 ------ACAGAUU--CUGAUGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGAUUUUAUAUGGAAGUCAGUUAGAGCCCCCCAGACUUGGGAGACG---------- ------.(((..(--(((..(.(((..(((....(((...((((..(....)..))))..))).(((((((.....))))))))))..))).)..)))).)))(....).---------- ( -26.20, z-score = -0.94, R) >droPer1.super_5 6335689 102 - 6813705 ------ACAGAUU--CUGAUGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGAUUUUAUAUGGAAGUCAGUUAGAGCCCCCCAGACUAGGGAGACG---------- ------......(--(((..(.(((..(((....(((...((((..(....)..))))..))).(((((((.....))))))))))..))).)..))))....(....).---------- ( -25.60, z-score = -0.90, R) >droMoj3.scaffold_6500 23360977 102 + 32352404 GCAGAGACACUUCAGCUGAUGAGCAUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGAUUUUAUAUGGAAGUCAAUAAUUUUGUUUGUUUUUG------------------ .((((((((...(((((....))).(((((((.(....).)))))))..(((((((.(((...(((....))).)))..)))))))....))..))))))))------------------ ( -24.90, z-score = -0.96, R) >droVir3.scaffold_12963 373300 101 - 20206255 GCAGAAACAGCUCAGCUGAUGAGCAUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGAUUUUAUAUGGAAGUCAAUAAUUU-GUUUUUGUUUG------------------ ((((((((((((((.....))))).(((((((.(....).)))))))..(((((((.(((...(((....))).)))..)))))))...)-))))))))...------------------ ( -31.80, z-score = -3.05, R) >consensus _______________ACAGAGAGCUUCAACACCGCUGGCAGUGUUGAAAUAUUGACACCACAGGUGAUUUUACAUGGAAGUCAAGAGGUUGAUUUGCCUUAGACUUAGGAAC_U______ ........................((((((((.(....).)))))))).......((((...))))...................................................... (-12.29 = -12.49 + 0.20)

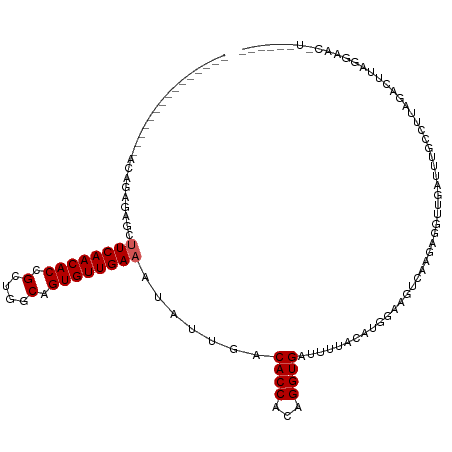
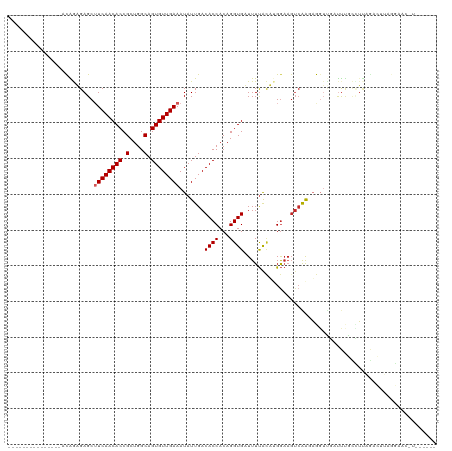
| Location | 5,438,639 – 5,438,738 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.42 |
| Shannon entropy | 0.57857 |
| G+C content | 0.43803 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -16.30 |
| Energy contribution | -16.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.998578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5438639 99 - 23011544 ------AGGUUCCUAAGUCUAAGGCAAAUCAACCUCUUGACUUCCAAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCUCUGU--------------- ------.((.....(((((..(((........)))...)))))))........(((((...))))).....(((((((((((....)))))))))))........--------------- ( -24.90, z-score = -1.76, R) >droSim1.chr2L 5241406 94 - 22036055 ------AGGUUCCUAAGUCUAAGGCAAAUCAACCUCUU-----CCAAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCUCUGU--------------- ------(((((.....((.....)).....)))))...-----..........(((((...))))).....(((((((((((....)))))))))))........--------------- ( -21.50, z-score = -1.38, R) >droSec1.super_5 3512950 99 - 5866729 ------AGGUUCCUAAGUCUAAGGCAAAUCAACCUCUUGACUUCCAAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCUCUGU--------------- ------.((.....(((((..(((........)))...)))))))........(((((...))))).....(((((((((((....)))))))))))........--------------- ( -24.90, z-score = -1.76, R) >droYak2.chr2L 8565599 99 + 22324452 ------AUGUUCCUAAGUCUAAGGCAAAUCAACCUCUUGACUUCCCAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCUCUGU--------------- ------........(((((..(((........)))...)))))..(((.....(((((...))))).....(((((((((((....)))))))))))....))).--------------- ( -25.20, z-score = -2.37, R) >droEre2.scaffold_4929 5522691 99 - 26641161 ------AGGUUCCUAAGUCUAAGGCAAAUCAACCUCUCGACUUCCCAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCUCUGU--------------- ------(((((.....((.....)).....)))))...(((..(((((.........))).)).)))....(((((((((((....)))))))))))........--------------- ( -24.50, z-score = -1.78, R) >droAna3.scaffold_12916 6465593 105 + 16180835 AGGUUAAAGACUUGGGUUUUUAAGCCAACUGACCUCUCGACUUUCCUCCGAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCUCUGU--------------- .((((((((((....)))))).))))...((((...(((.........))).((((....))))))))...(((((((((((....)))))))))))........--------------- ( -26.90, z-score = -0.96, R) >dp4.chr4_group1 4739982 102 - 5278887 ----------CGUCUCCCAAGUCUGGGGGGCUCUAACUGACUUCCAUAUAAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCAUCAG--AAUCUGU------ ----------.((((((((....))))))))(((...((((..(((((..........))))).))))...(((((((((((....)))))))))))......))--)......------ ( -35.00, z-score = -3.73, R) >droPer1.super_5 6335689 102 + 6813705 ----------CGUCUCCCUAGUCUGGGGGGCUCUAACUGACUUCCAUAUAAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCAUCAG--AAUCUGU------ ----------.(((((((......)))))))(((...((((..(((((..........))))).))))...(((((((((((....)))))))))))......))--)......------ ( -34.60, z-score = -3.62, R) >droMoj3.scaffold_6500 23360977 102 - 32352404 ------------------CAAAAACAAACAAAAUUAUUGACUUCCAUAUAAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAUGCUCAUCAGCUGAAGUGUCUCUGC ------------------................(((((((..(((((..........))))).))))))).....(((((..((((((((........)))).)))).)))))...... ( -24.90, z-score = -2.30, R) >droVir3.scaffold_12963 373300 101 + 20206255 ------------------CAAACAAAAAC-AAAUUAUUGACUUCCAUAUAAAAUCACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAUGCUCAUCAGCUGAGCUGUUUCUGC ------------------....((.((((-(...(((((((..(((((..........))))).)))))))..(((((((((....))))))))).(((((.....)))))))))).)). ( -31.90, z-score = -4.41, R) >consensus ______A_GUUCCUAAGUCUAAGGCAAAUCAACCUCUUGACUUCCAAGUAAAAACACCUGUGGUGUCAAUAUUUCAACACUGCCAGCGGUGUUGAAGCUCUCUGU_______________ ......................................................((((...))))......(((((((((((....)))))))))))....................... (-16.30 = -16.50 + 0.20)
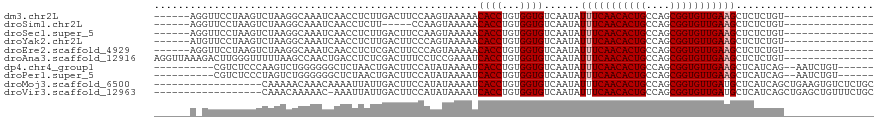

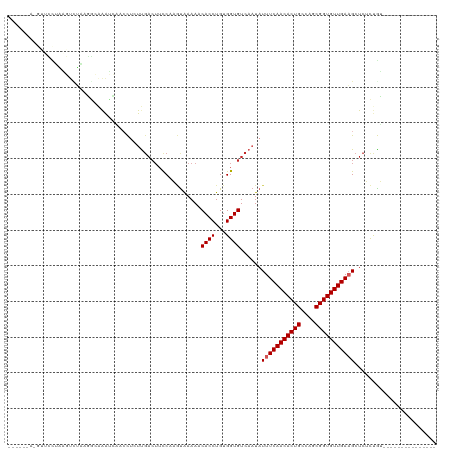
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:59 2011