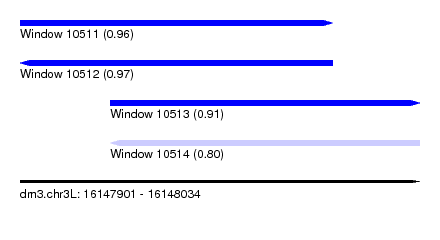
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,147,901 – 16,148,034 |
| Length | 133 |
| Max. P | 0.969409 |

| Location | 16,147,901 – 16,148,005 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.61 |
| Shannon entropy | 0.52448 |
| G+C content | 0.48010 |
| Mean single sequence MFE | -32.54 |
| Consensus MFE | -16.01 |
| Energy contribution | -16.54 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
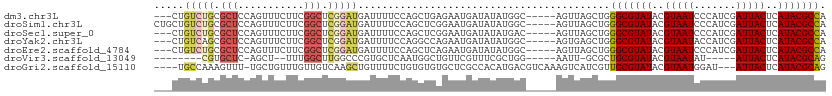
>dm3.chr3L 16147901 104 + 24543557 ---CUGUCUGCGCUCCAGUUUCUUCGGCUCGGAUGAUUUUCCAGCUGAGAAUGAUAUAUGGC-----AGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCA ---(((.((((........((((.(((((.(((......)))))))))))).........))-----)).)))...(((((((..((((((.....))))))..))))))). ( -35.63, z-score = -2.76, R) >droSim1.chr3L 15481138 107 + 22553184 CUGCUGUCUGCGCUCCAGUUUCUUCGGCUCGGAUGAUUUUCCAGCUCGGAAUGAUAUAUGGC-----AGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCA ..((((.((((.(....(((..(((((((.(((......)))))).))))..)))....)))-----)).))))..(((((((..((((((.....))))))..))))))). ( -38.70, z-score = -3.20, R) >droSec1.super_0 8247471 104 + 21120651 ---CUGUCUGCGCUCCAGUUUCUUCGGCUCGGAUGAUUUUCCAGCUCGGAAUGAUAUAUGAC-----AGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCA ---(((((.........(((((...((((.(((......))))))).))))).......)))-----)).......(((((((..((((((.....))))))..))))))). ( -31.99, z-score = -2.05, R) >droYak2.chr3L 2172365 104 - 24197627 ---CUGUCAGCGCUCCAGUUUCUUCGGCUCGGAUGAUUUUCCAGGCCAGAAUGAUAUAUGGC-----AGUGAGCUGGGCGUAUACGUAAUACCAUCGAUUACUCAUACGCCA ---....((((((((((...((((((((..(((......)))..))).))).))....))).-----)))..))))(((((((..(((((.......)))))..))))))). ( -37.30, z-score = -3.02, R) >droEre2.scaffold_4784 18395253 104 + 25762168 ---CUGUCUGCGCUCCAGUUUCUUCGGCUCGGAUGAUUUUCCAGCUCAGAAUGAUAUAUGGC-----AGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCA ---(((.((((........((((..((((.(((......))))))).)))).........))-----)).)))...(((((((..((((((.....))))))..))))))). ( -32.23, z-score = -2.04, R) >droVir3.scaffold_13049 1119505 90 + 25233164 --------CGUGCUC-AGCU--UUUGGCUUGGCCCGUGCUCAAUGGCUGUUCGUUUCGCUGG-----AAUU-GCGCUGCGUAUACGUAAUAU-----AUUACUCAUACGCAG --------.((((.(-((((--.(((((.........)).))).)))))((((......)))-----)...-))))(((((((..((((...-----.))))..))))))). ( -25.10, z-score = -0.21, R) >droGri2.scaffold_15110 2258626 104 + 24565398 ----UGCCAAAGUUU-UGCUGUUUGUUGUCAAGCUGUUUUCUGUGUGUGCUCGCCACAUGACGUCAAAGUCAUCGUUGCGUAUACGUAAUGGAU---AUUACUCAUACGCAG ----...........-.((.(((((....))))).))...(((((((((..(((.(((((((......))))).)).))).....(((((....---))))).))))))))) ( -26.80, z-score = -0.68, R) >consensus ___CUGUCUGCGCUCCAGUUUCUUCGGCUCGGAUGAUUUUCCAGCUCAGAAUGAUAUAUGGC_____AGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCA .....(((((.(((...........))).)))))..........................................(((((((..(((((.......)))))..))))))). (-16.01 = -16.54 + 0.53)


| Location | 16,147,901 – 16,148,005 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 72.61 |
| Shannon entropy | 0.52448 |
| G+C content | 0.48010 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -15.83 |
| Energy contribution | -16.04 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16147901 104 - 24543557 UGGCGUAUGAGUAAUCGAUGGGAUUACGUAUACGCCCAGCUAACU-----GCCAUAUAUCAUUCUCAGCUGGAAAAUCAUCCGAGCCGAAGAAACUGGAGCGCAGACAG--- .((((((((.((((((.....)))))).)))))))).......((-----((.........(((((.((((((......))).))).).))))........))))....--- ( -31.23, z-score = -2.09, R) >droSim1.chr3L 15481138 107 - 22553184 UGGCGUAUGAGUAAUCGAUGGGAUUACGUAUACGCCCAGCUAACU-----GCCAUAUAUCAUUCCGAGCUGGAAAAUCAUCCGAGCCGAAGAAACUGGAGCGCAGACAGCAG .((((((((.((((((.....)))))).))))))))..(((..((-----((.........(((((.((((((......))).)))))..)))........))))..))).. ( -33.73, z-score = -2.44, R) >droSec1.super_0 8247471 104 - 21120651 UGGCGUAUGAGUAAUCGAUGGGAUUACGUAUACGCCCAGCUAACU-----GUCAUAUAUCAUUCCGAGCUGGAAAAUCAUCCGAGCCGAAGAAACUGGAGCGCAGACAG--- .((((((((.((((((.....)))))).)))))))).......((-----(((.....((.(((((.((((((......))).)))))..)))....)).....)))))--- ( -31.80, z-score = -2.25, R) >droYak2.chr3L 2172365 104 + 24197627 UGGCGUAUGAGUAAUCGAUGGUAUUACGUAUACGCCCAGCUCACU-----GCCAUAUAUCAUUCUGGCCUGGAAAAUCAUCCGAGCCGAAGAAACUGGAGCGCUGACAG--- .((((((((.((((((....).))))).))))))))((((...((-----.(((....((.(((.(((.((((......)))).))))))))...))))).))))....--- ( -33.00, z-score = -2.20, R) >droEre2.scaffold_4784 18395253 104 - 25762168 UGGCGUAUGAGUAAUCGAUGGGAUUACGUAUACGCCCAGCUAACU-----GCCAUAUAUCAUUCUGAGCUGGAAAAUCAUCCGAGCCGAAGAAACUGGAGCGCAGACAG--- .((((((((.((((((.....)))))).)))))))).......((-----((.........((((..((((((......))).)))...))))........))))....--- ( -31.13, z-score = -2.06, R) >droVir3.scaffold_13049 1119505 90 - 25233164 CUGCGUAUGAGUAAU-----AUAUUACGUAUACGCAGCGC-AAUU-----CCAGCGAAACGAACAGCCAUUGAGCACGGGCCAAGCCAAA--AGCU-GAGCACG-------- (((((((((.((((.-----...)))).)))))))))(((-....-----...)))....(..((((..(((.((.........))))).--.)))-)..)...-------- ( -25.30, z-score = -1.73, R) >droGri2.scaffold_15110 2258626 104 - 24565398 CUGCGUAUGAGUAAU---AUCCAUUACGUAUACGCAACGAUGACUUUGACGUCAUGUGGCGAGCACACACAGAAAACAGCUUGACAACAAACAGCA-AAACUUUGGCA---- .((((((((.(((((---....))))).))))))))...(((((......)))))((..(((((..............)))))...)).....((.-........)).---- ( -23.64, z-score = -0.75, R) >consensus UGGCGUAUGAGUAAUCGAUGGGAUUACGUAUACGCCCAGCUAACU_____GCCAUAUAUCAUUCUGAGCUGGAAAAUCAUCCGAGCCGAAGAAACUGGAGCGCAGACAG___ .((((((((.(((((.......))))).))))))))..(((............................((((......))))..(((.......))))))........... (-15.83 = -16.04 + 0.21)

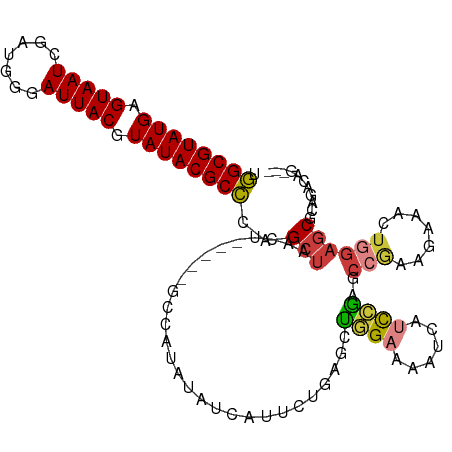
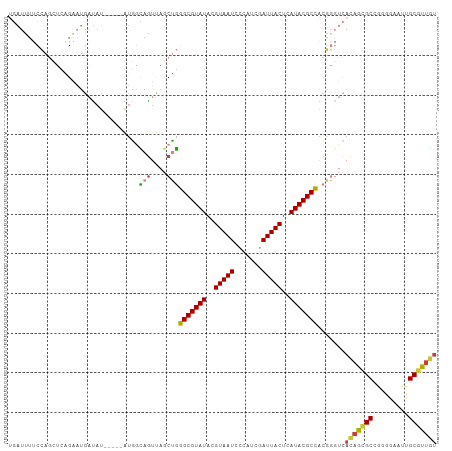
| Location | 16,147,931 – 16,148,034 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 73.78 |
| Shannon entropy | 0.50476 |
| G+C content | 0.47512 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -18.37 |
| Energy contribution | -17.94 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.909573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16147931 103 + 24543557 UGAUUUUCCAGCUGAGAAUGAUAU-----AUGGCAGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGGGUCACAGAGCCGUGGAAUUGCGUUGU .......((((((((...((....-----....)).))))))))((((((..((((((.....))))))..)))).((((((.((...)).))))))....))..... ( -33.70, z-score = -1.95, R) >droSim1.chr3L 15481171 103 + 22553184 UGAUUUUCCAGCUCGGAAUGAUAU-----AUGGCAGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGGGUCACAGCGCCGGCGAAUUGCGUUGU .....((((.....))))......-----...((((((.((..(((((((..((((((.....))))))..))))))).(((((....)).))))).))))))..... ( -32.40, z-score = -0.86, R) >droSec1.super_0 8247501 103 + 21120651 UGAUUUUCCAGCUCGGAAUGAUAU-----AUGACAGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGGGUCACAGCGCCGGCGAAUUGCGUUGU ..........(((((.........-----....(((....)))(((((((..((((((.....))))))..))))))).))))).(((((((.........))))))) ( -32.10, z-score = -1.21, R) >droYak2.chr3L 2172395 103 - 24197627 UGAUUUUCCAGGCCAGAAUGAUAU-----AUGGCAGUGAGCUGGGCGUAUACGUAAUACCAUCGAUUACUCAUACGCCACGGGUCACAGCGCCGGGGAAUUGCGUUGU .((((..((..((((.........-----.)))).((((.((((((((((..(((((.......)))))..))))))).))).))))......))..))))....... ( -37.40, z-score = -2.42, R) >droEre2.scaffold_4784 18395283 103 + 25762168 UGAUUUUCCAGCUCAGAAUGAUAU-----AUGGCAGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGGGUCACAGCGCCAAGGAAUUGCGUUGU .(((((..(((((.....((....-----....))...)))))(((((((..((((((.....))))))..)))))))..)))))(((((((.((....))))))))) ( -31.50, z-score = -1.71, R) >droVir3.scaffold_13049 1119527 91 + 25233164 CCGUGCUCAAUGGCUGUUCGUUUC-----GCUGGAAUUG-CGCUGCGUAUACGUAAU---AU--AUUACUCAUACGCAGACA---ACAAUGC---GAAAUUGCGUUUU ((((.....))))..((..(((((-----((.....(((-..((((((((..((((.---..--.))))..)))))))).))---)....))---))))).))..... ( -24.80, z-score = -0.83, R) >droGri2.scaffold_15110 2258654 100 + 24565398 CUGUUUUCUGUGUGUGCUCGCCACAUGACGUCAAAGUCAUCGUUGCGUAUACGUAAUGG-AU--AUUACUCAUACGCAGCAAU--AAAAUGC---GAAAUUGCGUCUC .........(((((.(....))))))((((.(((..((.....(((((((..(((((..-..--)))))..)))))))(((..--....)))---))..))))))).. ( -26.90, z-score = -1.40, R) >consensus UGAUUUUCCAGCUCAGAAUGAUAU_____AUGGCAGUUAGCUGGGCGUAUACGUAAUCCCAUCGAUUACUCAUACGCCACGGGUCACAGCGCCGGGGAAUUGCGUUGU .................................(((....)))(((((((..(((((.......)))))..))))))).......(((((((.........))))))) (-18.37 = -17.94 + -0.42)


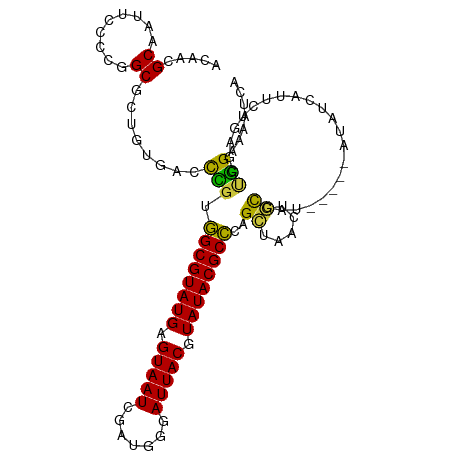
| Location | 16,147,931 – 16,148,034 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 73.78 |
| Shannon entropy | 0.50476 |
| G+C content | 0.47512 |
| Mean single sequence MFE | -29.26 |
| Consensus MFE | -15.53 |
| Energy contribution | -15.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.797228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16147931 103 - 24543557 ACAACGCAAUUCCACGGCUCUGUGACCCGUGGCGUAUGAGUAAUCGAUGGGAUUACGUAUACGCCCAGCUAACUGCCAU-----AUAUCAUUCUCAGCUGGAAAAUCA ............(((((.((...)).)))))(((((((.((((((.....)))))).)))))))((((((.........-----...........))))))....... ( -29.65, z-score = -1.72, R) >droSim1.chr3L 15481171 103 - 22553184 ACAACGCAAUUCGCCGGCGCUGUGACCCGUGGCGUAUGAGUAAUCGAUGGGAUUACGUAUACGCCCAGCUAACUGCCAU-----AUAUCAUUCCGAGCUGGAAAAUCA ........(((..(((((...((((...(.((((((((.((((((.....)))))).))))))))).((.....))...-----...)))).....)))))..))).. ( -31.00, z-score = -1.02, R) >droSec1.super_0 8247501 103 - 21120651 ACAACGCAAUUCGCCGGCGCUGUGACCCGUGGCGUAUGAGUAAUCGAUGGGAUUACGUAUACGCCCAGCUAACUGUCAU-----AUAUCAUUCCGAGCUGGAAAAUCA ........(((..(((((..((((((..(.((((((((.((((((.....)))))).)))))))))((....)))))))-----)..((.....)))))))..))).. ( -30.60, z-score = -0.92, R) >droYak2.chr3L 2172395 103 + 24197627 ACAACGCAAUUCCCCGGCGCUGUGACCCGUGGCGUAUGAGUAAUCGAUGGUAUUACGUAUACGCCCAGCUCACUGCCAU-----AUAUCAUUCUGGCCUGGAAAAUCA .........((((..(((((.((((.(.(.((((((((.((((((....).))))).))))))))).).)))).))((.-----(......).))))).))))..... ( -29.00, z-score = -0.82, R) >droEre2.scaffold_4784 18395283 103 - 25762168 ACAACGCAAUUCCUUGGCGCUGUGACCCGUGGCGUAUGAGUAAUCGAUGGGAUUACGUAUACGCCCAGCUAACUGCCAU-----AUAUCAUUCUGAGCUGGAAAAUCA .........((((..(((.(.((((...(.((((((((.((((((.....)))))).))))))))).((.....))...-----...))))...).)))))))..... ( -28.90, z-score = -1.04, R) >droVir3.scaffold_13049 1119527 91 - 25233164 AAAACGCAAUUUC---GCAUUGU---UGUCUGCGUAUGAGUAAU--AU---AUUACGUAUACGCAGCG-CAAUUCCAGC-----GAAACGAACAGCCAUUGAGCACGG .....((..((((---((...((---((((((((((((.((((.--..---.)))).))))))))).)-))))....))-----))))(((.......))).)).... ( -27.20, z-score = -2.26, R) >droGri2.scaffold_15110 2258654 100 - 24565398 GAGACGCAAUUUC---GCAUUUU--AUUGCUGCGUAUGAGUAAU--AU-CCAUUACGUAUACGCAACGAUGACUUUGACGUCAUGUGGCGAGCACACACAGAAAACAG ..(((((((..((---(((....--..)))((((((((.(((((--..-..))))).)))))))).....))..))).)))).((((.......)))).......... ( -28.50, z-score = -2.48, R) >consensus ACAACGCAAUUCCCCGGCGCUGUGACCCGUGGCGUAUGAGUAAUCGAUGGGAUUACGUAUACGCCCAGCUAACUGCCAU_____AUAUCAUUCUGAGCUGGAAAAUCA ...............(((...((.......((((((((.(((((.......))))).))))))))......)).)))............................... (-15.53 = -15.53 + 0.00)

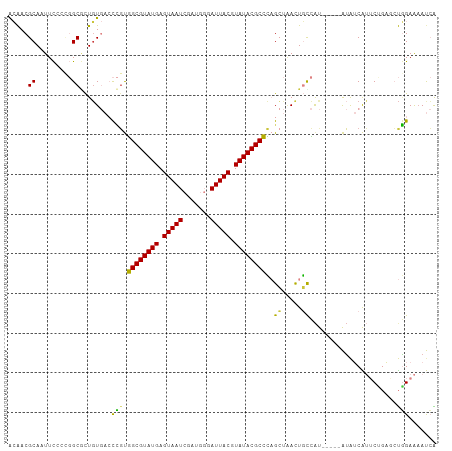
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:40 2011