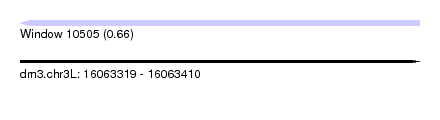
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,063,319 – 16,063,410 |
| Length | 91 |
| Max. P | 0.661851 |

| Location | 16,063,319 – 16,063,410 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 63.47 |
| Shannon entropy | 0.82671 |
| G+C content | 0.47315 |
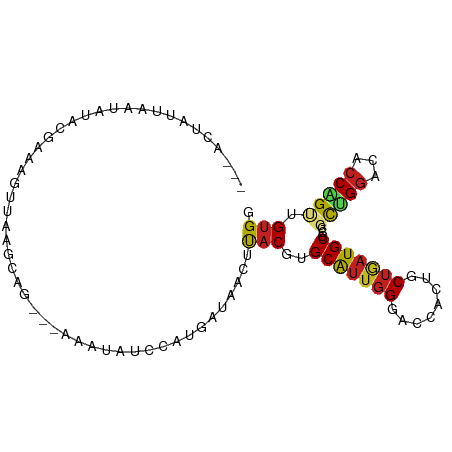
| Mean single sequence MFE | -22.39 |
| Consensus MFE | -10.53 |
| Energy contribution | -10.68 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.47 |
| Mean z-score | -0.28 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.661851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16063319 91 - 24543557 AUUACUAUU--UAUUCAAAAGUUGGGCAG----AAUAUCAUUGAUAACUUACGUGCGAUGGGACCGCUGCUGAUGCGCCGCUGGACACCAGUUGUGG .........--...........(.(((((----....((((((...((....)).)))))).....))))).)..(((.(((((...))))).))). ( -21.80, z-score = -0.12, R) >droGri2.scaffold_15110 5020162 93 - 24565398 ---UCGAUUAACAAAACUAAAAUUGU-AUAUUUUCAACAUAAGCCAACUUACGCGCAUUGGGUCCACUGCUGAUGCGACGCUGCGAGCCCGCUGUGC ---.......((((........))))-............((((....)))).(((((.((((..(.(.((.........)).).)..)))).))))) ( -16.90, z-score = 1.02, R) >droMoj3.scaffold_6680 18664799 89 + 24764193 ----CGUUUAAUA----UGAGCUCAUCACAGUUACAUUUGCAGUCAACUUACGCGCAUUGGGCCCACUGCUGAUGCGACGCUGGCUGCCCGCGGUGC ----(((.....(----((((((......)))).)))..(((((((.....(((((((..(........)..))))).)).)))))))..))).... ( -23.10, z-score = 1.27, R) >droVir3.scaffold_13049 9249384 89 - 25233164 --------UAAUGUAAAUAAAGGUUUUCCUUAACUGUUUCAAGUCAACUUACGCGCAUUGGGUCCACUGCUUAUGCGACGCGGGUUGCCCGUUGUGC --------....((((...((((....)))).(((......)))....))))(((((.(((((...((((.........))))...))))).))))) ( -23.60, z-score = -0.80, R) >droWil1.scaffold_181024 858022 91 - 1751709 ------AUUAAAGAGUUUGCCAGAAGUUUUUAAUUACAGUUAUUUUACUUACGGGCAUUCGGACCACUGCUAAUGCGACGCUGGACACCAGUUGUGG ------........(..((((..((((...((((....))))....))))...))))..)...(((((((....)))..(((((...))))).)))) ( -21.50, z-score = -1.16, R) >droPer1.super_9 3576272 91 + 3637205 ---ACCAUUUGAAUGGGGAAUCAUAGGAG---GACUCAUCAUAAAUACUCACGGGCAUUGGGACCACUGCUGAUGCGUCGCUGGAUACCAGUUGUGG ---.((((.....(((...(((...((..---(((.((((.......(....)((((.((....)).)))))))).))).)).))).)))...)))) ( -21.90, z-score = 0.49, R) >dp4.chrXR_group6 11546476 90 + 13314419 ---ACCAUUUGAAUGGGGAAUCAUAGGAG---GACUCAUCAU-AAUACUCACGGGCAUUGGGACCACUGCUGAUGCGUCGCUGGAUACCAGUUGUGG ---.((((.....(((...(((...((..---(((.((((..-....(....)((((.((....)).)))))))).))).)).))).)))...)))) ( -21.90, z-score = 0.55, R) >droAna3.scaffold_13337 6740362 89 + 23293914 ----CCAUUCAAAUAUAAAUCUUAAACA----GAAAUUGAGAGAAGACUUACGUGCAUUGGGACCACUGCUGAUGCGCCUUUGGACUCCGGUUGCGG ----...((((((......((((((...----....))))))..........((((((..(........)..)))))).))))))..(((....))) ( -17.80, z-score = -0.08, R) >droEre2.scaffold_4784 18310019 91 - 25762168 AUUACUAUUA--AUCCGAAAGUUAGGCG----GAAUAUACAUGAUAACUUACGUGCAUUGGGACCGCUGCUGAUGCGCCUCUGGACACCAGUUGUGG ..........--.((((.........))----)).....((..(........((((((..(........)..))))))..((((...)))))..)). ( -19.10, z-score = 0.47, R) >droYak2.chr3L 18628645 97 - 24197627 AUUACUAUUACUAUCUGGAAGUUAGGCGCGUCUAAUAUCCAUGAUAACUCACGUGCAUUGGGACCGCUGCUGAUGCGCCUCUGGACACCAGUCGUGG ...........((((((((.((((((....)))))).)))).))))...(((((((((..(........)..))))))..((((...))))..))). ( -29.20, z-score = -1.50, R) >droSec1.super_0 8163189 93 - 21120651 AUUACUAUU--UAUUCGAAAGUUGAGCAGA--AAAUAUCCUUGAUAACUUACGUGCAUUGGGACCGCUGCUGAUGCGCCGCUGGACACCAGUGGUGG .........--....(....).(.(((((.--.....((((.....((....)).....))))...))))).)..(((((((((...))))))))). ( -25.30, z-score = -1.44, R) >droSim1.chr3L 15403473 93 - 22553184 AUUACUAUU--UAUACGAAAGUUGAGCAGA--AAAUAUCCUUGAUAACUUACGUGCAUUGGGACCGCUGCUGAUGCGCCGCUGGACACCAGUGGUGG .........--...((....))(.(((((.--.....((((.....((....)).....))))...))))).)..(((((((((...))))))))). ( -26.60, z-score = -2.07, R) >consensus ___ACUAUUAAUAUACGAAAGUUAAGCAG___AAAUAUCCAUGAUAACUUACGUGCAUUGGGACCACUGCUGAUGCGCCGCUGGACACCAGUUGUGG .................................................(((..(((((((........)))))))...(((((...))))).))). (-10.53 = -10.68 + 0.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:33 2011