
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,063,182 – 16,063,279 |
| Length | 97 |
| Max. P | 0.719207 |

| Location | 16,063,182 – 16,063,279 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 70.21 |
| Shannon entropy | 0.61567 |
| G+C content | 0.44569 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -12.27 |
| Energy contribution | -12.18 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.719207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
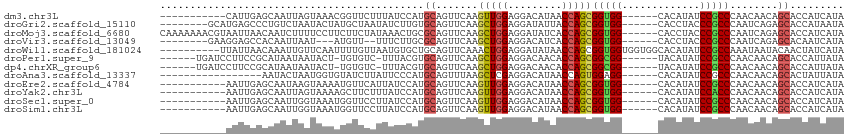
>dm3.chr3L 16063182 97 + 24543557 -----------CAUUGAGCAAUUAGUAAACGGUUCUUUAUCCAUGCAGUUCAAGUUGGAGGACAUAACCAGCGGUGG------CACAUAUCCGCCCAACAACAGCACCAUCAUA -----------..((((((.....(....)((........)).....))))))(((((.((......)).((((...------.......)))))))))............... ( -19.00, z-score = -0.17, R) >droGri2.scaffold_15110 5020022 100 + 24565398 --------GCAUGAGCCCUGUCUAAUACUAUGCUAAUAUCUUGUGCAGUUCAAGCUGGAGGAUAUUACCAGCGGUGG------CACCUACCCGCCCAAUCAGAGCACCAUAAUA --------(((..((....((.((....)).))......))..))).((((..(((((.........)))))(((((------.......)))))......))))......... ( -24.60, z-score = -0.14, R) >droMoj3.scaffold_6680 18664651 108 - 24764193 CAAAAAAACGUAAUUAACAAUCUUUUCCUUCUUCUAUAAACUGCGCAGUUCAAGCUGGAGGAUAUCACCAGCGGUGG------CACCUACCCGCCCAAUCAGAGCACCAUCAUA ...............................................((((..(((((..(....).)))))(((((------.......)))))......))))......... ( -19.30, z-score = -0.59, R) >droVir3.scaffold_13049 9249249 95 + 25233164 --------GAAGGAGCCACAAUUAAU---AUGUU--UUUCUUGCGCAGUUCAAGCUGGAGGACAUCACCAGCGGUGG------CACCUACCCGCCCAAUCAGAGCACAAUCAUA --------(((((.(((((.......---(((((--((((..((.........)).)))))))))........))))------).)))....((.(.....).))....))... ( -23.16, z-score = -0.69, R) >droWil1.scaffold_181024 857878 104 + 1751709 ----------UUAUUAACAAAUUGUUCAAUUUUGUUAAUGUGCUGCAGUUCAAACUGGAGGAUAUAACCAGCGGUGGUGGUGGCACAUAUCCGCCAAAUAAUACAACUAUCAUA ----------.((((((((((.........))))))))))..((.((((....)))).))((((..((((....))))(((((.......)))))............))))... ( -23.60, z-score = -0.71, R) >droPer1.super_9 3576132 100 - 3637205 ------UGAUCCUUCCGCAUAAUAAUACU-UGUGUC-UUUACGUGCAGUUCAAGCUGGAGGACAACACCAGCGGCGG------UACAUAUCCGCCCAACAACAGCACCAUUAUA ------..........((((((......)-))))).-.....((((.(((...(((((.(.....).)))))(((((------.......)))))....))).))))....... ( -27.10, z-score = -2.26, R) >dp4.chrXR_group6 11546336 100 - 13314419 ------UGAUCCUUCCGCAUAAUAAUACU-UGUGUC-UUUACGUGCAGUUCAAGCUGGAGGACAACACCAGCGGCGG------UACAUAUCCGCCCAACAACAGCACCAUUAUA ------..........((((((......)-))))).-.....((((.(((...(((((.(.....).)))))(((((------.......)))))....))).))))....... ( -27.10, z-score = -2.26, R) >droAna3.scaffold_13337 6740231 91 - 23293914 -----------------AAUACUAAUGGUGUAUCUUAUUCCCAUGCAGUUUAAGCUCGAGGACAUAACCAGUGGAGG------CACAUAUCCGCCCAACAACAGCACUAUUAUA -----------------.....(((((((((...((((((((..((.......))..).))).))))...(((((..------......))))).........))))))))).. ( -19.30, z-score = -0.97, R) >droEre2.scaffold_4784 18309882 97 + 25762168 -----------AAUUGAGCAAUAAGUAAAAUGUUCAUUAUCCAUGCAGUUCAAGUUGGAGGACAUAACCAGCGGUGG------CACAUAUCCGCCCAACAACAGCACCAUCAUA -----------...((((((..........)))))).......(((.(((...(((((.((......)).((((...------.......)))))))))))).)))........ ( -19.20, z-score = -0.57, R) >droYak2.chr3L 18628508 97 + 24197627 -----------AAUUGAGCAAUUAGUAAAAGCUUCUUUAUCCAUGCAGUUCAAGUUGGAGGACAUAACCAGCGGUGG------CACAUAUCCACCCAACAACAGCACCAUCAUA -----------..((((((.((..(((((......)))))..))...))))))((((..((......)).(.(((((------.......))))))..))))............ ( -17.30, z-score = -0.19, R) >droSec1.super_0 8163052 97 + 21120651 -----------AAUUGAGCAAUUGGUAAAUGGUUCCUUAUCCAUGCAGUUCAAGUUGGAGGACAUAACCAGCGGUGG------CACAUAUCCGCCCAACAACAGCACCAUCAUA -----------...........((((..((((........))))((.(((...(((((.((......)).((((...------.......)))))))))))).))))))..... ( -21.40, z-score = 0.15, R) >droSim1.chr3L 15403336 97 + 22553184 -----------AAUUGAGCAAUUGGUAAAUGGUUCCUUAUCCAUGCAGUUCAAGUUGGAGGACAUAACCAGCGGUGG------CACAUAUCCGCCCAACAACAGCACCAUCAUA -----------...........((((..((((........))))((.(((...(((((.((......)).((((...------.......)))))))))))).))))))..... ( -21.40, z-score = 0.15, R) >consensus ___________AAUUGACCAAUUAAUAAAAUGUUCCUUAUCCAUGCAGUUCAAGCUGGAGGACAUAACCAGCGGUGG______CACAUAUCCGCCCAACAACAGCACCAUCAUA ............................................((.......(((((.........)))))(((((.............)))))........))......... (-12.27 = -12.18 + -0.09)

| Location | 16,063,182 – 16,063,279 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 70.21 |
| Shannon entropy | 0.61567 |
| G+C content | 0.44569 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -12.81 |
| Energy contribution | -12.79 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.610157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
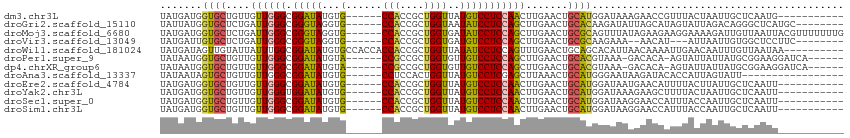
>dm3.chr3L 16063182 97 - 24543557 UAUGAUGGUGCUGUUGUUGGGCGGAUAUGUG------CCACCGCUGGUUAUGUCCUCCAACUUGAACUGCAUGGAUAAAGAACCGUUUACUAAUUGCUCAAUG----------- ..(((..((((.(((((((((.(((((((.(------(((....)))))))))))))))))...))).))))((........)).............)))...----------- ( -25.40, z-score = -0.97, R) >droGri2.scaffold_15110 5020022 100 - 24565398 UAUUAUGGUGCUCUGAUUGGGCGGGUAGGUG------CCACCGCUGGUAAUAUCCUCCAGCUUGAACUGCACAAGAUAUUAGCAUAGUAUUAGACAGGGCUCAUGC-------- ...(((((.((((((.(((.((((...((..------...))(((((.........))))).....)))).)))(((((((...)))))))...))))))))))).-------- ( -30.00, z-score = -0.21, R) >droMoj3.scaffold_6680 18664651 108 + 24764193 UAUGAUGGUGCUCUGAUUGGGCGGGUAGGUG------CCACCGCUGGUGAUAUCCUCCAGCUUGAACUGCGCAGUUUAUAGAAGAAGGAAAAGAUUGUUAAUUACGUUUUUUUG (((((..((((...(.(..(((.((..((((------.((((...)))).))))..)).)))..).).))))...))))).......((((((((.((.....)))))))))). ( -27.80, z-score = -0.30, R) >droVir3.scaffold_13049 9249249 95 - 25233164 UAUGAUUGUGCUCUGAUUGGGCGGGUAGGUG------CCACCGCUGGUGAUGUCCUCCAGCUUGAACUGCGCAAGAAA--AACAU---AUUAAUUGUGGCUCCUUC-------- ......(.(((((.....))))).).(((.(------((((.(((((.(.....).)))))........(....)...--.....---.......))))).)))..-------- ( -27.70, z-score = -0.75, R) >droWil1.scaffold_181024 857878 104 - 1751709 UAUGAUAGUUGUAUUAUUUGGCGGAUAUGUGCCACCACCACCGCUGGUUAUAUCCUCCAGUUUGAACUGCAGCACAUUAACAAAAUUGAACAAUUUGUUAAUAA---------- .......(((((((((.((((.(((((((.((((.(......).))))))))))).))))..)))..))))))..(((((((((.........)))))))))..---------- ( -26.90, z-score = -2.38, R) >droPer1.super_9 3576132 100 + 3637205 UAUAAUGGUGCUGUUGUUGGGCGGAUAUGUA------CCGCCGCUGGUGUUGUCCUCCAGCUUGAACUGCACGUAAA-GACACA-AGUAUUAUUAUGCGGAAGGAUCA------ ((((((((((((..((((.(((((.......------)))))(((((.(.....).)))))................-))))..-))))))))))))(....).....------ ( -31.50, z-score = -1.24, R) >dp4.chrXR_group6 11546336 100 + 13314419 UAUAAUGGUGCUGUUGUUGGGCGGAUAUGUA------CCGCCGCUGGUGUUGUCCUCCAGCUUGAACUGCACGUAAA-GACACA-AGUAUUAUUAUGCGGAAGGAUCA------ ((((((((((((..((((.(((((.......------)))))(((((.(.....).)))))................-))))..-))))))))))))(....).....------ ( -31.50, z-score = -1.24, R) >droAna3.scaffold_13337 6740231 91 + 23293914 UAUAAUAGUGCUGUUGUUGGGCGGAUAUGUG------CCUCCACUGGUUAUGUCCUCGAGCUUAAACUGCAUGGGAAUAAGAUACACCAUUAGUAUU----------------- ......((((((((..(((((((((((((.(------((......))))))))))....))))))...))((((............)))).))))))----------------- ( -21.10, z-score = 0.22, R) >droEre2.scaffold_4784 18309882 97 - 25762168 UAUGAUGGUGCUGUUGUUGGGCGGAUAUGUG------CCACCGCUGGUUAUGUCCUCCAACUUGAACUGCAUGGAUAAUGAACAUUUUACUUAUUGCUCAAUU----------- .......((((.(((((((((.(((((((.(------(((....)))))))))))))))))...))).))))(..((((((.........))))))..)....----------- ( -26.20, z-score = -1.45, R) >droYak2.chr3L 18628508 97 - 24197627 UAUGAUGGUGCUGUUGUUGGGUGGAUAUGUG------CCACCGCUGGUUAUGUCCUCCAACUUGAACUGCAUGGAUAAAGAAGCUUUUACUAAUUGCUCAAUU----------- .....(.((((.(((((((((.(((((((.(------(((....)))))))))))))))))...))).)))).).....((.((...........))))....----------- ( -26.00, z-score = -1.34, R) >droSec1.super_0 8163052 97 - 21120651 UAUGAUGGUGCUGUUGUUGGGCGGAUAUGUG------CCACCGCUGGUUAUGUCCUCCAACUUGAACUGCAUGGAUAAGGAACCAUUUACCAAUUGCUCAAUU----------- .....((((((.(((((((((.(((((((.(------(((....)))))))))))))))))...))).))((((........))))..))))...........----------- ( -27.70, z-score = -0.94, R) >droSim1.chr3L 15403336 97 - 22553184 UAUGAUGGUGCUGUUGUUGGGCGGAUAUGUG------CCACCGCUGGUUAUGUCCUCCAACUUGAACUGCAUGGAUAAGGAACCAUUUACCAAUUGCUCAAUU----------- .....((((((.(((((((((.(((((((.(------(((....)))))))))))))))))...))).))((((........))))..))))...........----------- ( -27.70, z-score = -0.94, R) >consensus UAUGAUGGUGCUGUUGUUGGGCGGAUAUGUG______CCACCGCUGGUUAUGUCCUCCAGCUUGAACUGCAUGGAAAAAAAACAAUUUACUAAUUGCUCAAUU___________ .......((((....(((((((((................)))).((......)).))))).......)))).......................................... (-12.81 = -12.79 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:32 2011