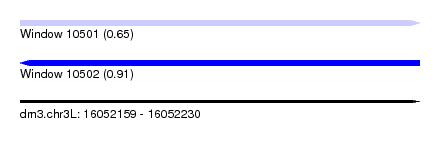
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,052,159 – 16,052,230 |
| Length | 71 |
| Max. P | 0.912055 |

| Location | 16,052,159 – 16,052,230 |
|---|---|
| Length | 71 |
| Sequences | 6 |
| Columns | 71 |
| Reading direction | forward |
| Mean pairwise identity | 87.22 |
| Shannon entropy | 0.24361 |
| G+C content | 0.35053 |
| Mean single sequence MFE | -18.10 |
| Consensus MFE | -12.25 |
| Energy contribution | -13.50 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.653226 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
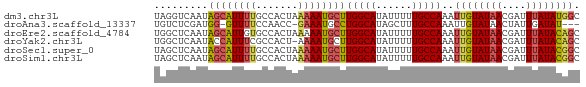
>dm3.chr3L 16052159 71 + 24543557 GCCAUAUAAAUCGUUAUACAAUUUGGCAAAAAUAUGCCAAGCAUUUUUAGUGGCAAAAUGCUAUUGACCUA ((..(((((....)))))....((((((......))))))))....((((((((.....)))))))).... ( -15.50, z-score = -1.40, R) >droAna3.scaffold_13337 6730866 66 - 23293914 ---AUAUCAAUAGUUAUACAAUUUGGCAAAGCUAUGCCAGGCAUUUC-GGUUGGAAAAC-CCAUCGAGACA ---..................(((((((......)))))))..((((-(((.((....)-).))))))).. ( -14.80, z-score = -1.59, R) >droEre2.scaffold_4784 18298346 71 + 25762168 GCUGUAUAAAUCGUUAUACAAUUUGGCAAAAAUAUGCCAAGCAUUUUUAGUGGCACAAUGCUAUUGAGCCA (((((((((....)))))))..((((((......))))))))...(((((((((.....)))))))))... ( -20.90, z-score = -2.53, R) >droYak2.chr3L 18614167 70 + 24197627 GCUGUAUAAAUCGUUAUACAAUUUGGCAAAAAUAUGCCAAGCAUUUU-AGUGGCGAAAUGGUAUUGAGCCA (((((((((....)))))))..((((((......)))))))).....-..((((..(((...)))..)))) ( -17.40, z-score = -1.45, R) >droSec1.super_0 8152238 71 + 21120651 GCCGUAUAAAUCGUUAUACAAUUUGGCAAAAAUAUGCCAAGCAUUUUUAGUGGCAAAAUGCUAUUGAGCUA ((.((((((....))))))...((((((......))))))))...(((((((((.....)))))))))... ( -20.00, z-score = -2.52, R) >droSim1.chr3L 15390766 71 + 22553184 GCCGUAUAAAUCGUUAUACAAUUUGGCAAAAAUAUGCCAAGCAUUUUUAGUGGCAAAAUGCUAUUGAGCUA ((.((((((....))))))...((((((......))))))))...(((((((((.....)))))))))... ( -20.00, z-score = -2.52, R) >consensus GCCGUAUAAAUCGUUAUACAAUUUGGCAAAAAUAUGCCAAGCAUUUUUAGUGGCAAAAUGCUAUUGAGCCA ...((((((....))))))..(((((((......)))))))....(((((((((.....)))))))))... (-12.25 = -13.50 + 1.25)

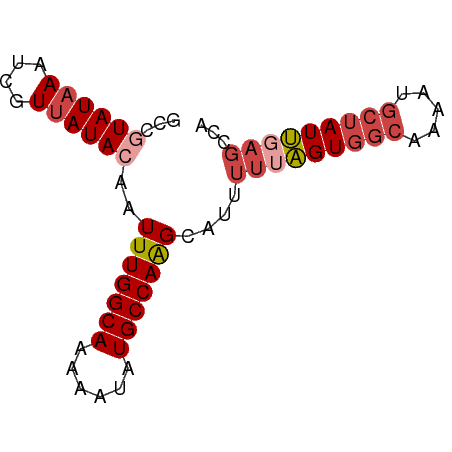
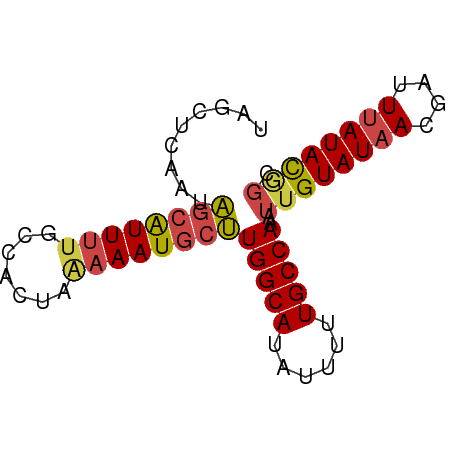
| Location | 16,052,159 – 16,052,230 |
|---|---|
| Length | 71 |
| Sequences | 6 |
| Columns | 71 |
| Reading direction | reverse |
| Mean pairwise identity | 87.22 |
| Shannon entropy | 0.24361 |
| G+C content | 0.35053 |
| Mean single sequence MFE | -16.99 |
| Consensus MFE | -13.83 |
| Energy contribution | -14.08 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912055 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 16052159 71 - 24543557 UAGGUCAAUAGCAUUUUGCCACUAAAAAUGCUUGGCAUAUUUUUGCCAAAUUGUAUAACGAUUUAUAUGGC ((((((....(((((((.......)))))))((((((......))))))..........))))))...... ( -17.80, z-score = -1.98, R) >droAna3.scaffold_13337 6730866 66 + 23293914 UGUCUCGAUGG-GUUUUCCAACC-GAAAUGCCUGGCAUAGCUUUGCCAAAUUGUAUAACUAUUGAUAU--- ....(((((((-...(((.....-)))((((.(((((......)))))....))))..)))))))...--- ( -13.50, z-score = -0.84, R) >droEre2.scaffold_4784 18298346 71 - 25762168 UGGCUCAAUAGCAUUGUGCCACUAAAAAUGCUUGGCAUAUUUUUGCCAAAUUGUAUAACGAUUUAUACAGC ((((.((((...)))).))))..........((((((......)))))).((((((((....)))))))). ( -19.30, z-score = -2.22, R) >droYak2.chr3L 18614167 70 - 24197627 UGGCUCAAUACCAUUUCGCCACU-AAAAUGCUUGGCAUAUUUUUGCCAAAUUGUAUAACGAUUUAUACAGC ((((.............))))..-.......((((((......)))))).((((((((....)))))))). ( -15.72, z-score = -2.21, R) >droSec1.super_0 8152238 71 - 21120651 UAGCUCAAUAGCAUUUUGCCACUAAAAAUGCUUGGCAUAUUUUUGCCAAAUUGUAUAACGAUUUAUACGGC ..(((.....(((((((.......)))))))((((((......))))))...((((((....))))))))) ( -17.80, z-score = -2.28, R) >droSim1.chr3L 15390766 71 - 22553184 UAGCUCAAUAGCAUUUUGCCACUAAAAAUGCUUGGCAUAUUUUUGCCAAAUUGUAUAACGAUUUAUACGGC ..(((.....(((((((.......)))))))((((((......))))))...((((((....))))))))) ( -17.80, z-score = -2.28, R) >consensus UAGCUCAAUAGCAUUUUGCCACUAAAAAUGCUUGGCAUAUUUUUGCCAAAUUGUAUAACGAUUUAUACGGC .........((((((((.......))))))))(((((......)))))..((((((((....)))))))). (-13.83 = -14.08 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:30 2011