| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,429,624 – 5,429,723 |
| Length | 99 |
| Max. P | 0.500000 |

| Location | 5,429,624 – 5,429,723 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.04 |
| Shannon entropy | 0.31832 |
| G+C content | 0.43708 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -17.66 |
| Energy contribution | -17.28 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
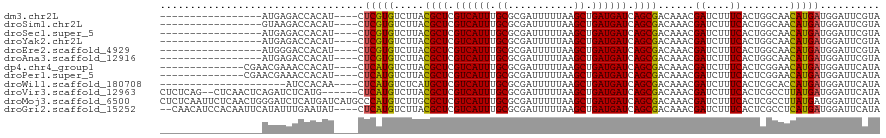
>dm3.chr2L 5429624 99 + 23011544 -----------------AUGAGACCACAU----CUCGUGUCUUACGCUCGUCAUUUGCGCGAUUUUUAAGCUGAUGAUCAGCGACAAACGAUCUUUCACUGGCAACAUGAUGGAUUCGUA -----------------.((((((.((..----...))))))))((((.((((((.((...........)).)))))).))))....(((((((.(((.((....))))).))).)))). ( -30.50, z-score = -2.31, R) >droSim1.chr2L 5232518 99 + 22036055 -----------------GUAAGACCACAU----CUCGUGUCUUACGCUCGUCAUUUGCGCGAUUUUUAAGCUGAUGAUCAGCGACAAACGAUCUUUCACUGGCAACAUGAUGGAUUCGUA -----------------(((((((.((..----...)))))))))(((.((((((.((...........)).)))))).))).....(((((((.(((.((....))))).))).)))). ( -32.00, z-score = -3.05, R) >droSec1.super_5 3504070 99 + 5866729 -----------------AUGAGACCACAU----CUCGUGUCUUACGCUCGUCAUUUGCGCGAUUUUUAAGCUGAUGAUCAGCGACAAACGAUCUUUCACUGGCAACAUGAUGGAUUCGUA -----------------.((((((.((..----...))))))))((((.((((((.((...........)).)))))).))))....(((((((.(((.((....))))).))).)))). ( -30.50, z-score = -2.31, R) >droYak2.chr2L 8556235 99 - 22324452 -----------------AUGAGACCACAU----CUCGUGUCUUACGCUCGUCAUUUGCGCGAUUUUUAAGCUGAUGAUCAGCGACAAACGAUCUUUCACUGGCAACAUGAUGGAUUCGUA -----------------.((((((.((..----...))))))))((((.((((((.((...........)).)))))).))))....(((((((.(((.((....))))).))).)))). ( -30.50, z-score = -2.31, R) >droEre2.scaffold_4929 5511915 99 + 26641161 -----------------AUGGGACCACAU----CUCGUGUCUUACGCUCGUCAUUUGCGCGAUUUUUAAGCUGAUGAUCAGCGACAAACGAUCUUUCACUGGCAACAUGAUGGAUUCGUA -----------------.((((((.((..----...))))))))((((.((((((.((...........)).)))))).))))....(((((((.(((.((....))))).))).)))). ( -29.70, z-score = -2.02, R) >droAna3.scaffold_12916 6456473 99 - 16180835 -----------------AUGAGACCACAU----CUCGUGUCUUACGCUCGUCAUUUGCGCGAUUUUUAAGCUGAUGAUCAGCGACAAACGAUCUUUCACUGGCAACAUGAUGGAUUCGUA -----------------.((((((.((..----...))))))))((((.((((((.((...........)).)))))).))))....(((((((.(((.((....))))).))).)))). ( -30.50, z-score = -2.31, R) >dp4.chr4_group1 4730353 102 + 5278887 --------------CGAACGAAACCACAU----CUCAUGUCUUACGCUCGUCAUUUGCGCGAUUUUUAAGCUGAUGAUCAGCGACAAACGAUCUUUCACUCGGAACAUGAUGGAUUCAUA --------------.....(((.(((...----.((((((....((((.((((((.((...........)).)))))).)))).....(((........)))..))))))))).)))... ( -25.50, z-score = -1.59, R) >droPer1.super_5 6326016 102 - 6813705 --------------CGAACGAAACCACAU----CUCAUGUCUUACGCUCGUCAUUUGCGCGAUUUUUAAGCUGAUGAUCAGCGACAAACGAUCUUUCACUCGGAACAUGAUGGAUUCAUA --------------.....(((.(((...----.((((((....((((.((((((.((...........)).)))))).)))).....(((........)))..))))))))).)))... ( -25.50, z-score = -1.59, R) >droWil1.scaffold_180708 572496 95 + 12563649 ---------------------AUCCACAA----CUCAUGUCUCAUGCUCGUCAUUUGCGCGAUUUUUAAGCUGAUGAUCAGCGACAAACGAUCUUUCACUCGCACCAUGAUGGAUUCAUA ---------------------(((((...----.(((((......((.........))((((.......((((.....)))).......((....))..))))..))))))))))..... ( -20.30, z-score = -0.63, R) >droVir3.scaffold_12963 359717 112 - 20206255 CUCUCAG--CUCAACUCAGAUCUCAUG------CUCAUGUCUUACGCUCGUCAUUUGCGCGAUUUUUAAGCUGAUGAUCAGCGACAAACGAUCUUUCACUCGCCUUAUGAUGGAUUCAUA ......(--(.......(((((.(((.------...))).....((((.((((((.((...........)).)))))).))))......))))).......))..(((((.....))))) ( -21.34, z-score = 0.55, R) >droMoj3.scaffold_6500 23346349 120 + 32352404 CUCUCAAUUCUCAACUGGGAUCUCAUGAUCAUGCCCAUGUCUUGCGCUCGUCAUUUGCGCGAUUUUUAAGCUGAUGAUCAGCGACAAACGAUCUUUCACUCGCCUUAUGAUGGAUUCAUA .....((((((((...((((((....)))).......((((((((((.........)))))).......((((.....))))))))................))...))).))))).... ( -27.60, z-score = -0.35, R) >droGri2.scaffold_15252 5878369 114 - 17193109 --CAACAUCCACAAUUCAUAUUUGAAUAU----CUCAUGUCUUACGCUCGUCAUUUGCGCGAUUUUUAAGCUGAUGAUCAGCGACAAACGAUCUUUCACUCGCCUCAUGAUGGAUUCAUA --....(((((..(((((....)))))..----.(((((.....((((.((((((.((...........)).)))))).)))).....(((........)))...))))))))))..... ( -25.40, z-score = -1.74, R) >consensus _________________AUGAGACCACAU____CUCAUGUCUUACGCUCGUCAUUUGCGCGAUUUUUAAGCUGAUGAUCAGCGACAAACGAUCUUUCACUCGCAACAUGAUGGAUUCAUA ..................................(((((.....((((.((((((.((...........)).)))))).))))......((....))........))))).......... (-17.66 = -17.28 + -0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:54 2011