
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,038,285 – 16,038,379 |
| Length | 94 |
| Max. P | 0.875361 |

| Location | 16,038,285 – 16,038,379 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 61.02 |
| Shannon entropy | 0.77352 |
| G+C content | 0.50644 |
| Mean single sequence MFE | -27.61 |
| Consensus MFE | -9.27 |
| Energy contribution | -9.09 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16038285 94 - 24543557 AUCGUCAUCGGAGGCACCACCAAUUCCGGACUAUGGAGCAUUAGCUUCUUUG--UGGAACAUCCGCUGCCAACGGAAGUGGUGCACUCAUGCGUGU----- ..((.(((..((((((((((...(((((......(((((....)))))...(--(((.....))))......)))))))))))).))))))))...----- ( -34.70, z-score = -2.21, R) >droEre2.scaffold_4784 18283922 96 - 25762168 GGCGUCAUCGCCAGUACCACCAAUUCCGGACUAUGGAGCAUUAGCUUCUUUGUCUGGCACAUCCGCUGCCAGCGGAAGUGGCGCACUCAUGCGUGU----- ((((....)))).............((((((...(((((....)))))...))))))(((.(((((.....))))).)))(((((....)))))..----- ( -40.00, z-score = -3.01, R) >droSec1.super_0 8138682 88 - 21120651 ------AUCGUCAGCACCACCAAUUCCGGACUAUGGAGCAUUAGCUUCUUUG--UGGCACAUCCGCUGCCAACGGAAGUGGCGCACUCAAGCGUGU----- ------.......((.((((...(((((.((...(((((....)))))...)--)((((.......))))..))))))))).))............----- ( -27.00, z-score = -0.76, R) >droSim1.chr3L 15376393 88 - 22553184 ------AUCGUCAGCACCACCAAUUCCGGACUAUGGAGCAUUAGCUUCUUUG--UGGCACAUCCGCUGCCAACGGAAGUGGUGCACUCAUGCGUGU----- ------.......(((((((...(((((.((...(((((....)))))...)--)((((.......))))..))))))))))))............----- ( -30.90, z-score = -1.85, R) >droPer1.super_9 3545837 87 + 3637205 ------CUUCUCCUCUCUGUCACUCAC---CAGCAAAGCAUUACCUUCUUUGUCUGGCACAUCCGCUCUGAACGGAAGUGGUGCACUCAUGCGUGC----- ------....................(---((((((((.(......)))))).))))(((.((((.......)))).)))..((((......))))----- ( -20.00, z-score = -0.51, R) >dp4.chrXR_group6 11517376 87 + 13314419 ------CUUCUCCUCUCUGUCACUCAC---CAGCAAAGCAUUACCUUCUUUGUCUGGCACAUCCGCUCUGAACGGAAGUGGUGCACUCAUGCGUGC----- ------....................(---((((((((.(......)))))).))))(((.((((.......)))).)))..((((......))))----- ( -20.00, z-score = -0.51, R) >droWil1.scaffold_181024 818405 94 - 1751709 --AAGGCAAAAGCAAUGUUUUUUGUUUUUGUUUUUUGGGCCCUGUUUCUUUGUCUAGCAUUUCCGUUGUGAACGGAAGUGGUGCACUCAUGCGUGU----- --(((((((((((((......)))))))))))))((((((...........))))))(((((((((.....)))))))))..((((......))))----- ( -26.80, z-score = -1.86, R) >droMoj3.scaffold_6680 18640383 90 + 24764193 ------CUGCAAAGAAGUAUUUAGAGUUUUUCUUUU-----UCUUUGCUUUGUUCCUCAUUUCCGUUUGCAGCGGAAGUGGUUCGCUCAUGCGUGCGCCGC ------..(((((((((.....((((...)))).))-----))))))).........(((((((((.....)))))))))((.(((....))).))..... ( -26.70, z-score = -2.25, R) >droVir3.scaffold_13049 9223144 92 - 25233164 ---AGACCGAGCAUAUGUAUUU-GAGUAAUUUUUUU-----UGUUUGCUUUGUUCGUAAUUUCCGCUCGCAACGGAAGUGGUGCACUCAUGCGUACGCCGC ---....((((((...(((..(-(((........))-----))..)))..))))))....(((((.......)))))((((((.((......)).)))))) ( -22.40, z-score = -0.19, R) >consensus ______CUCGUCAGCACUAUCAAUUCCGGACUAUGGAGCAUUAGCUUCUUUGUCUGGCACAUCCGCUGCCAACGGAAGUGGUGCACUCAUGCGUGU_____ .......................................................(.(((.((((.......)))).))).)((((......))))..... ( -9.27 = -9.09 + -0.18)

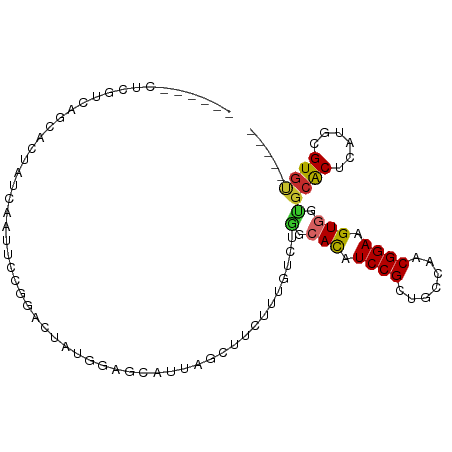
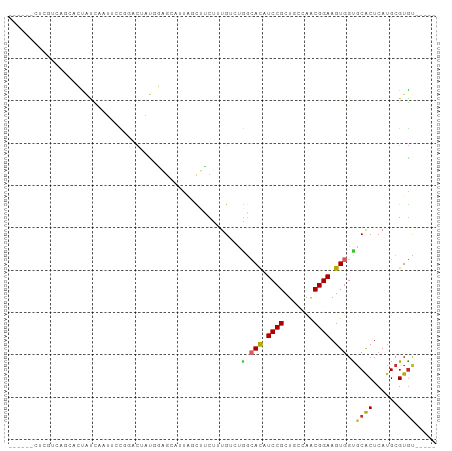
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:28 2011