
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,030,195 – 16,030,280 |
| Length | 85 |
| Max. P | 0.582899 |

| Location | 16,030,195 – 16,030,280 |
|---|---|
| Length | 85 |
| Sequences | 11 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 56.04 |
| Shannon entropy | 0.84554 |
| G+C content | 0.36449 |
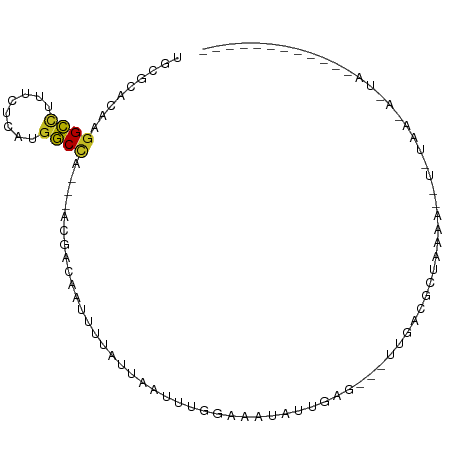
| Mean single sequence MFE | -14.48 |
| Consensus MFE | -5.58 |
| Energy contribution | -5.26 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.75 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.582899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
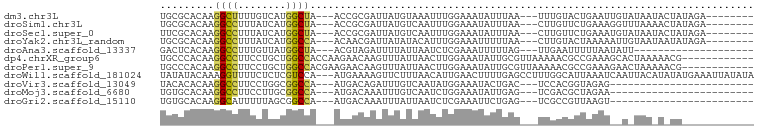
>dm3.chr3L 16030195 85 + 24543557 UGCGCACAAGGCUUUUGUCAUGGCUA---ACCGCGAUUAUGUAAAUUUGGAAAUAUUUAA---UUUGUACUGAAUUGUAUAAUACUAUAGA-------- ..(((....(((....)))..((...---.))))).((((((((((.(....).))))).---.((((((......))))))...))))).-------- ( -11.70, z-score = 0.98, R) >droSim1.chr3L 15368279 85 + 22553184 UGCGCACAAGGCCUUUAUCAUGGCUA---ACCGCGAUUAUGUCAAUUUGGAAAUAUUUAA---CUUGUUCUGAAAGGUUUAAAACUAUAGA-------- ..(((....((((........)))).---...)))..........((..(((.((.....---..)))))..)).................-------- ( -11.40, z-score = 1.13, R) >droSec1.super_0 8130684 85 + 21120651 UUCGCACAAGGCCUUUAUCAUGGCUA---ACCGCGAUUAUGUCAAUUUGGAAAUAUUUAA---CUUGUUCUGAAAUGUAUAAUACUAUAGA-------- .((((....((((........)))).---...))))(((((.((.((..(((.((.....---..)))))..)).))))))).........-------- ( -16.00, z-score = -1.20, R) >droYak2.chr3L_random 3179171 85 - 4797643 UGCGCACAAGGCCUUUAUCAUGGCCA---ACAACGAUUAUAUACAUUUGGAAAUUUUUAA---CUUGUACUAAAAAUUGUAAUAAUAUAGA-------- .........((((........)))).---...((((((...((((.(((((....)))))---..)))).....))))))...........-------- ( -12.20, z-score = -0.42, R) >droAna3.scaffold_13337 6708980 73 - 23293914 GACUCACAAGGCCUUUGUUAUGGCUA---ACGUAGAUUUUAUUAAUCUCGAAAUUUUUAG---UUGAAUUUUUAAUAUU-------------------- ((((.....((((........)))).---.((.(((((.....)))))))........))---))..............-------------------- ( -11.00, z-score = -1.02, R) >dp4.chrXR_group6 11508879 87 - 13314419 UGCCCACAAGGCCUUCCUGCUGGCCACCAAGAACAAGUUUAUUAACUUGGAAAUAUUGCGUUAAAAACGCCGAAAGCACUAAAAACG------------ (((......((((........))))........((((((....))))))........((((.....)))).....))).........------------ ( -18.50, z-score = -1.41, R) >droPer1.super_9 3537141 87 - 3637205 UGCCCACAAGGCCUUCCUGCUGGCCACGAAGAACAAGUUUAUUAACUUGGAAAUAUUGCGUUAAAAACGCCGAAAGAACUAAAAACG------------ .........((((........))))........((((((....))))))........((((.....))))(....)...........------------ ( -18.00, z-score = -1.52, R) >droWil1.scaffold_181024 802772 96 + 1751709 UAUAUACAAAGGUUUUCUCUCGUCCA---AUGAAAAGUUCUUUAACAUUGAACUUUUGAGCCUUUGGCAUUAAAUCAAUUACAUAUAUGAAAUUAUAUA ......(((((((((...........---...((((((((.........)))))))))))))))))................(((((......))))). ( -16.13, z-score = -1.81, R) >droVir3.scaffold_13049 9213244 69 + 25233164 UACACACAAGGCCUUCCUGGCGGCCA---AUGACAGAUUUGUCAAUAUGGAAAUACUGAC---UCCACGGUAGAG------------------------ .........((((........)))).---.((((......)))).........(((((..---....)))))...------------------------ ( -14.70, z-score = -0.52, R) >droMoj3.scaffold_6680 18630934 69 - 24764193 UGUGCACAAGGCCUUCCUUGCGGCCA---AUGACAAAUUUGUCAAUCUGGAAAUAUUGAG---UCGACGCUAGAA------------------------ .((((((..((((........)))).---.(((((....)))))...............)---).).))).....------------------------ ( -14.30, z-score = 0.03, R) >droGri2.scaffold_15110 4984822 69 + 24565398 UGUGCACAAGGCAUUUUUAGCGGCCA---AUGACAAAUUUAUUAAUCUCGAAAUUCUGAG---UCGCCGUUAAGU------------------------ .((((.....)))).(((((((((.(---((((.....)))))...((((......))))---..))))))))).------------------------ ( -15.40, z-score = -1.29, R) >consensus UGCGCACAAGGCCUUUCUCAUGGCCA___ACGACAAUUUUAUUAAUUUGGAAAUAUUGAG___UUGACGCUAAAA__U_UAA_A_UA____________ .........((((........)))).......................................................................... ( -5.58 = -5.26 + -0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:27 2011