
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,014,406 – 16,014,510 |
| Length | 104 |
| Max. P | 0.500000 |

| Location | 16,014,406 – 16,014,510 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 88.38 |
| Shannon entropy | 0.23940 |
| G+C content | 0.50495 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -21.52 |
| Energy contribution | -22.49 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
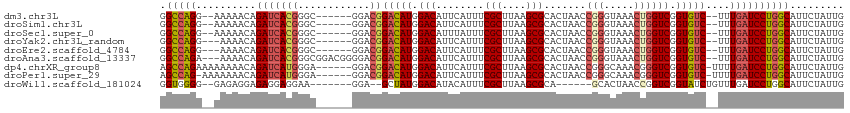
>dm3.chr3L 16014406 104 - 24543557 GGCCAGG--AAAAACAGAUCACGGGC------GGACGGACAUGGACAUUCAUUUCGCUUAAGCGCACUAACCGGGUAAACUGGUCGGUGUC--UUUGAUCCUGGCAUUCUAUUG .((((((--(............((((------(((..((.........))..)))))))(((.((((..(((((.....)))))..)))))--))...)))))))......... ( -34.70, z-score = -2.72, R) >droSim1.chr3L 15352487 104 - 22553184 GGCCAGG--AAAAACAGAUCACGGGC------GGACGGACAUGGACAUUCAUUUCGCUUAAGCGCACUAACCGGGUAAACUGGUCGGUGUC--UUUGAUCCUGGCAUUCUAUUG .((((((--(............((((------(((..((.........))..)))))))(((.((((..(((((.....)))))..)))))--))...)))))))......... ( -34.70, z-score = -2.72, R) >droSec1.super_0 8115013 104 - 21120651 GGCCAGG--AAAAACAGAUCACGGGC------GGACGGACAUGGACAUUUAUUUCGCUUAAGCGCACUAACCGGGUAAACUGGUCGGUGUC--UUUGAUCCUGGCAUUCUAUUG .((((((--(....((..((.((...------...))))..))..............(((((.((((..(((((.....)))))..)))).--))))))))))))......... ( -34.20, z-score = -2.74, R) >droYak2.chr3L_random 3154772 103 + 4797643 GGCCAGG---AAAACAGAUCACGGGC------GGACGGACAUGGACAUUCAUUUCGCUUAAGCGCACUAACCGGGUAAACUGGUCGGUGUC--UUUGAUCCUGGCAUUCUAUUG .((((((---(...........((((------(((..((.........))..)))))))(((.((((..(((((.....)))))..)))))--))...)))))))......... ( -34.70, z-score = -2.68, R) >droEre2.scaffold_4784 18259181 103 - 25762168 GGCCAGG---AAAACAGAUCACGGGC------GGACGGACAUGGACAUUCAUUUCGCUUAAGCGCACUAACCGGGUAAACUGGUCGGUGUC--UUUGAUCCUGGCAUUCUAUUG .((((((---(...........((((------(((..((.........))..)))))))(((.((((..(((((.....)))))..)))))--))...)))))))......... ( -34.70, z-score = -2.68, R) >droAna3.scaffold_13337 6690340 109 + 23293914 GGCCAGA---AAAACAGAUCACGGGCGGACGGGGACGGACAUGGACAUUCAUUUCGCUUAAGCGCACUAACCGGGUAAACUGGUCGGUGUC--UUUGAUCCUGGCAUUCUAUUG .(((((.---......(((((.(((((((((....))...((((....)))))))))))(((.((((..(((((.....)))))..)))))--))))))))))))......... ( -36.41, z-score = -2.44, R) >dp4.chrXR_group8 4258268 107 - 9212921 AGCCAGAAAAAAAACAGAUCAUGGGA------GGACGGACAUGGACAUUCAUUUCGCUUAAGCGCACUAACCGGGCAAACGGGUCGGUGUC-UUUUGAUCCUGGCAUUCUAUUG .(((((..........(((((..(((------((.((((.((((....)))))))))))....((((..(((.(.....).)))..)))))-)..))))))))))......... ( -30.00, z-score = -1.48, R) >droPer1.super_29 491313 106 - 1099123 AGCCAG-AAAAAAACAGAUCAUGGGA------GGACGGACAUGGACAUUCAUUUCGCUUAAGCGCACUAACCGGGCAAACGGGUCGGUGUC-UUUUGAUCCUGGCAUUCUAUUG .(((((-.........(((((..(((------((.((((.((((....)))))))))))....((((..(((.(.....).)))..)))))-)..))))))))))......... ( -30.10, z-score = -1.48, R) >droWil1.scaffold_181024 774061 97 - 1751709 GGUGGGG--GAGAGGAGAGGAGGAA-------GGA--GCUAUGGACAUACAUUUCGCUUAAGCGCA------GCACUAACCGGUCGGUAUCUGUUUGAUCCUGGCAUUCUAUUG (((((((--.....(..((((..((-------(.(--(.(((.(((........(((....)))..------..........))).))).)).)))..))))..).))))))). ( -20.45, z-score = 1.93, R) >consensus GGCCAGG__AAAAACAGAUCACGGGC______GGACGGACAUGGACAUUCAUUUCGCUUAAGCGCACUAACCGGGUAAACUGGUCGGUGUC__UUUGAUCCUGGCAUUCUAUUG .(((((..........(((((................((.(((......))).))((....))((((..(((((.....)))))..)))).....))))))))))......... (-21.52 = -22.49 + 0.97)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:26 2011