
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,988,742 – 15,988,843 |
| Length | 101 |
| Max. P | 0.524991 |

| Location | 15,988,742 – 15,988,843 |
|---|---|
| Length | 101 |
| Sequences | 15 |
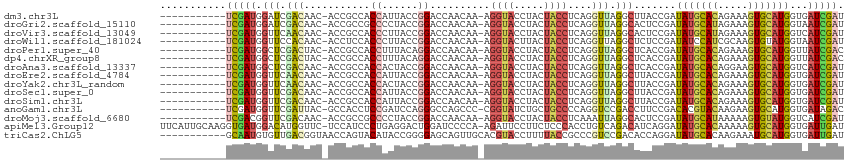
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.03 |
| Shannon entropy | 0.46657 |
| G+C content | 0.50481 |
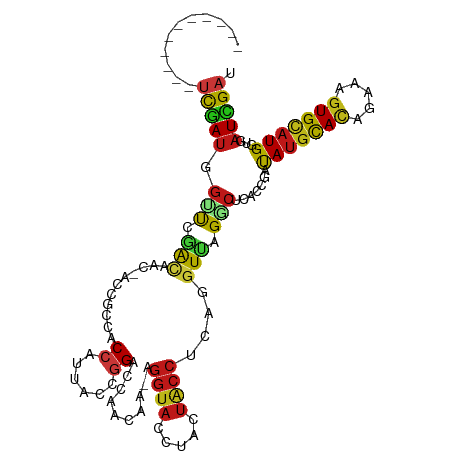
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -14.05 |
| Energy contribution | -14.31 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.524991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15988742 101 - 24543557 -----------UCGAUGGAUCGACAAC-ACCGCCACCAUUACCGGACCAACAA-AGGUACCUACUACCUCAGGUUAGGCUUACCGAUAUGCACAGAAAGUGCAUGGUGAUCGAU -----------.......(((((...(-((((((.((......))...(((..-(((((.....)))))...))).)))........((((((.....)))))))))).))))) ( -28.40, z-score = -2.17, R) >droGri2.scaffold_15110 8920169 101 + 24565398 -----------UCGAUGGAUCGACAAC-ACCGCCGCCCCUACCGGACCAACAA-AGGUACCUACUACCUCAGGUUAGGCACUCCGAUAUGCAUAGAAAGUGCAUGGUAAUCGAU -----------(((((((..((.(...-...).))..))((((((((((((..-(((((.....)))))...))).))...)))...((((((.....))))))))))))))). ( -25.90, z-score = -0.96, R) >droVir3.scaffold_13049 14588175 101 - 25233164 -----------UCGAUGGUUCAACAAC-ACCGCCACCCUUACCGGACCAACAA-AGGUACCUACUACCUCAGGUUAGGCACUCCGAUAUGCAUAGAAAGUGCAUGGUCAUCGAU -----------(((((((((....)))-...(((........(((((((((..-(((((.....)))))...))).))...))))..((((((.....))))))))))))))). ( -26.90, z-score = -1.73, R) >droWil1.scaffold_181024 1612263 101 - 1751709 -----------UCGAUGGUUCCACAAC-ACCUCCACCCUUACCGGACCAACAA-AGGUACUUACUACCUCAGGUUAGGCUCUCCGAUAUCCAUCGCAAGUGUAUGGUAAUCGAU -----------(((((....(((..((-((.........((.(((((((((..-(((((.....)))))...))).))...)))).))..........)))).)))..))))). ( -22.01, z-score = -0.36, R) >droPer1.super_40 380297 101 + 810552 -----------UCGAUGGCUCGACUAC-ACCGCCACCUUUACAGGACCAACAA-AGGUACCUACUACCUCAGGUUAGGCUCACCGAUAUGCACAGAAAGUGCAUGGUUAUCGAC -----------((((((((........-...(((.(((....)))...(((..-(((((.....)))))...))).))).......(((((((.....))))))))))))))). ( -29.50, z-score = -2.51, R) >dp4.chrXR_group8 973444 101 + 9212921 -----------UCGAUGGCUCGACUAC-ACCGCCACCUUUACAGGACCAACAA-AGGUACCUACUACCUCAGGUUAGGCUCACCGAUAUGCACAGAAAGUGCAUGGUUAUCGAC -----------((((((((........-...(((.(((....)))...(((..-(((((.....)))))...))).))).......(((((((.....))))))))))))))). ( -29.50, z-score = -2.51, R) >droAna3.scaffold_13337 12126900 101 + 23293914 -----------UCGAUGGCUCGACAAC-ACCGCCACCACUACCGGACCAACAA-AGGUACCUACUACCUCAGGUUAGGCUCACCGAUAUGCACAGGAAGUGCAUGGUCAUCGAU -----------((((((((........-...(((.((......))...(((..-(((((.....)))))...))).))).......(((((((.....))))))))))))))). ( -31.20, z-score = -2.39, R) >droEre2.scaffold_4784 18233015 101 - 25762168 -----------UCGAUGGUUCAACAAC-ACCGCCACCAUUACCGGACCAACAA-AGGUACCUACUACCUCAGGUUAGGCUUACCGAUAUGCACAGAAAGUGCAUGGUGAUCGAU -----------((((((((........-)))..((((.....(((.(((((..-(((((.....)))))...))).))....)))..((((((.....))))))))))))))). ( -29.50, z-score = -2.52, R) >droYak2.chr3L_random 3129022 101 + 4797643 -----------UCGAUGGUUCAACAAC-ACCGCCACCACUACCGGACCAACAA-AGGUACCUACUACCUCAGGUUAGGCUUACCGAUAUGCACAGAAAGUGCAUGGUGAUCGAU -----------((((((((........-)))..((((.....(((.(((((..-(((((.....)))))...))).))....)))..((((((.....))))))))))))))). ( -29.50, z-score = -2.44, R) >droSec1.super_0 8089011 101 - 21120651 -----------UCGAUGGUUCGACAAC-ACCGCCACCAUUACCGGACCAACAA-AGGUACCUACUACCUCAGGUUAGGCUUACCGAUAUGCACAGAAAGUGCAUGGUGAUCGAU -----------((((((((........-)))..((((.....(((.(((((..-(((((.....)))))...))).))....)))..((((((.....))))))))))))))). ( -29.50, z-score = -2.27, R) >droSim1.chr3L 15326316 101 - 22553184 -----------UCGAUGGUUCGACAAC-ACCGCCACCAUUACCGGACCAACAA-AGGUACCUACUACCUCAGGUUAGGCUUACCGAUAUGCACAGAAAGUGCAUGGUGAUCGAU -----------((((((((........-)))..((((.....(((.(((((..-(((((.....)))))...))).))....)))..((((((.....))))))))))))))). ( -29.50, z-score = -2.27, R) >anoGam1.chr3L 23566625 101 - 41284009 -----------UCGAUGGUUCGAUUAC-GCCACCUCCGAUCCAGGGCCAGCCC-CGGUAUCUGCUGCCCCAGGUCCGACCUUCCGACACGUACAAGAAGUGCAUGGUGAUAGAC -----------((((....))))...(-((((..((.((((..((((.(((..-........)))))))..)))).))...........((((.....)))).)))))...... ( -27.40, z-score = 0.54, R) >droMoj3.scaffold_6680 12052286 101 + 24764193 -----------UCGACGGUUCGACAAC-ACCGCCGCCCCUACCGGACCAACAA-AGGUACCUACUACCUCAAAUUAGGCACUCCGAUAUGCAUAAAAAGUGUAUGGUCAUCGAU -----------((((((((........-))))..(((((....))........-(((((.....))))).......))).....(((((((((.....)))))).))).)))). ( -20.90, z-score = -0.26, R) >apiMel3.Group12 3007793 112 + 9182753 UUCAUUGCAAGGUGAUGGACAUGGUUC-UCCAUCCCUGAGGACUGGAUCCCCA-AGAUUCCUUCUCCCACCUGUCAGACAUCAGGAUAUGCACAAAAAGUGCAUGGUGAUUGAU .((((..(.(((.((((((........-)))))))))(((((..(((((....-.))))).)))))...((((........)))).(((((((.....))))))))..).))). ( -33.80, z-score = -0.50, R) >triCas2.ChLG5 18511484 103 - 18847211 -----------GCAAUGUGUUGACGGUAACCAGUACAUACCGGGGAGCAGUUGCACGUACCUUUUACCGCCCGUCCGACACCAGGAUAUGCACAAGAAAUGCAUGGUGAUUGAU -----------(((((.((((..(((((.........)))))...)))))))))......(..(((((....((((.......))))(((((.......))))))))))..).. ( -26.10, z-score = 0.43, R) >consensus ___________UCGAUGGUUCGACAAC_ACCGCCACCAUUACCGGACCAACAA_AGGUACCUACUACCUCAGGUUAGGCUCACCGAUAUGCACAGAAAGUGCAUGGUGAUCGAU ...........(((((.(((.(((...........((......))..........((((.....))))....))).))).......(((((((.....)))))))...))))). (-14.05 = -14.31 + 0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:22 2011