
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,422,559 – 5,422,661 |
| Length | 102 |
| Max. P | 0.514180 |

| Location | 5,422,559 – 5,422,661 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.83 |
| Shannon entropy | 0.39568 |
| G+C content | 0.40547 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -15.06 |
| Energy contribution | -15.44 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5422559 102 + 23011544 CAAAGUGGCUUU-UGUGCUGGUGCUGCUGCUGCUGUUGUUGUCGACGCCAAUUCUGAUUAUUCACAUCGAUUGGAAUUUGUGAAA-UUUUCACGAAUUGACACU (((((..((...-.((((....)).)).))..)).))).((((....((((((.(((....)))....))))))(((((((((..-...))))))))))))).. ( -25.50, z-score = -0.81, R) >droSim1.chr2L 5215431 96 + 22036055 CAAAGUGGCUUU-UGGGCUG------CUGCUGCUGUUGUUGUCGCCGCCAAUUCUGAUUAUUCACAUCGAUUGGAAUUUGUGAAA-UUUUCACGAAUUGACACU ...((..(((..-..)))..------))...........((((....((((((.(((....)))....))))))(((((((((..-...))))))))))))).. ( -24.40, z-score = -0.96, R) >droSec1.super_5 3497107 93 + 5866729 CAAAGUGGCUUU-UGUGCUG---------CUGCCGUUGUUGUCGCCGCCAAUUCUGAUUAUUCACAUCGAUUGGAAUUUGUGAAA-UUUUCACGAAUUGACACU ...((..((...-...))..---------))........((((....((((((.(((....)))....))))))(((((((((..-...))))))))))))).. ( -23.90, z-score = -1.36, R) >droYak2.chr2L 8548891 99 - 22324452 CAAAGUGGCUUU-UGUGCUG---CUGUUGUUGCUGUUGUUGUCGCCGCCAAUUCUGAUUAUUCACAUCGAUUGGAAUUUGUGAAA-UUUUCACGAAUUGACACU (((((..((...-...))..---)).)))..........((((....((((((.(((....)))....))))))(((((((((..-...))))))))))))).. ( -24.10, z-score = -0.94, R) >droEre2.scaffold_4929 5504530 90 + 26641161 CAAAGUGGCUUU-UGUGCUG------------CUGUUGUUGUCGCCGCCAAUUCUGAUUAUUCACAUCGAUUGGAAUUUGUGAAA-UUUUCACGAAUUGACACU (((((..((...-...))..------------)).))).((((....((((((.(((....)))....))))))(((((((((..-...))))))))))))).. ( -24.10, z-score = -1.95, R) >droAna3.scaffold_12916 6447726 88 - 16180835 CAAAGUAGCAAUCCCAAUUG---------------UUAUUGUCGCCGCCAAUUCUGAUUAUUCACAUCGAUUGGAAUUUGUGAAA-UUUUCACGAAUUGACACU ...(((((((((....))))---------------)))))(((....((((((.(((....)))....))))))(((((((((..-...))))))))))))... ( -21.90, z-score = -2.86, R) >dp4.chr4_group1 4722909 86 + 5278887 CAAAG-AGCAGUCC-AAUUG---------------UUGUUGUCGCCGCCAAUUCUGAUUAUUCACAUCGAUUGGAAUCUGUGAAA-UUCUCACGAAUUGACACU ...((-(....(((-(((((---------------.(((.((((..........)))).....))).)))))))).)))(((.((-(((....)))))..))). ( -17.30, z-score = -0.16, R) >droPer1.super_5 6318592 86 - 6813705 CAAAG-AGCAGUCC-AAUUG---------------UUGUUGUCGCCGCCAAUUCUGAUUAUUCACAUCGAUUGGAAUCUGUGAAA-UUCUCACGAAUUGACACU ...((-(....(((-(((((---------------.(((.((((..........)))).....))).)))))))).)))(((.((-(((....)))))..))). ( -17.30, z-score = -0.16, R) >droWil1.scaffold_180708 560979 89 + 12563649 CAAAGUUGUUCU-U-UGUUG-------------CGUUUCUAUCGAUUCCGUUUCUGAUUAUUCACAUCGAUUGGAAUUUGUGAAAACUUUCACGAAUUGACACU (((((.....))-)-))...-------------........(((((((...(((..(((.........)))..)))...(((((....)))))))))))).... ( -16.20, z-score = -0.27, R) >droGri2.scaffold_15252 5868819 81 - 17193109 ----CUCACUCUCUCCGUUG-----------------GCUGCUGUUGUCGCUGCUGAUUAUUCACAUCGAUUGGCAUUUGUGAAA--UUUCACGAAUUGGCACU ----............((..-----------------(.((((...((((.((.(((....))))).)))).))))(((((((..--..))))))))..))... ( -18.50, z-score = -0.88, R) >consensus CAAAGUGGCUUU_UGUGCUG______________GUUGUUGUCGCCGCCAAUUCUGAUUAUUCACAUCGAUUGGAAUUUGUGAAA_UUUUCACGAAUUGACACU .......................................((((....((((((.(((....)))....))))))((((((((((....)))))))))))))).. (-15.06 = -15.44 + 0.38)
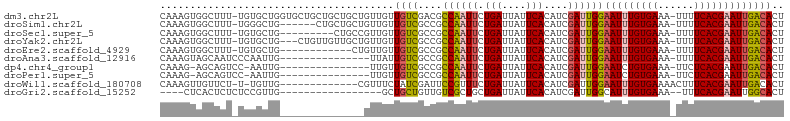
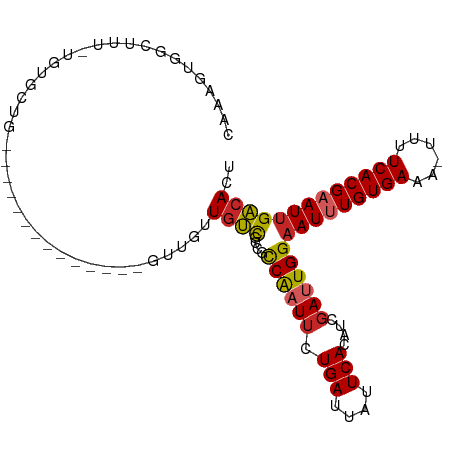

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:53 2011