| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,939,554 – 15,939,647 |
| Length | 93 |
| Max. P | 0.886887 |

| Location | 15,939,554 – 15,939,647 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 72.08 |
| Shannon entropy | 0.44454 |
| G+C content | 0.33763 |
| Mean single sequence MFE | -13.98 |
| Consensus MFE | -11.40 |
| Energy contribution | -11.28 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.886887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15939554 93 - 24543557 -------------------------CUCUCUUAAAAUAAAAUCAUUGUUUUACGAAACAUUUCCAAGAUUCCUCAAAGCAAUUAUCCACUUGCUGUCUGGACUGCUUCG-UUUGCAGUU -------------------------..................(((((...(((((.((..((((.(((.......(((((........)))))))))))).)).))))-)..))))). ( -16.91, z-score = -2.10, R) >droEre2.scaffold_4784 18184016 95 - 25762168 ---------------------UUUCAUAUAUAUACACAAAAA-AUCGAUUCACACAACAUUUCUAAGAUUCCUCAAAG-AAUUAGCCACUUGCUGUCUGGACUGCUUCG-UUUGCAGUA ---------------------.....................-......(((.(((.((.......(((((......)-)))).......)).))).)))(((((....-...))))). ( -11.94, z-score = 0.06, R) >droYak2.chr3L 18533204 116 - 24197627 AUUUAUACUAUUUUACCCAUAUCUUAAGUAUAUAAAU-GAAU-AUCGAUUUAAAAAACAUUUCUAACAUUCCUCGAAGCAAUUAUCCACUUGCUGUCUGGACUGCUUCG-UUUGCAGUA ................(((.((..(((((..((((.(-(...-.((((........................))))..)).))))..)))))..)).)))(((((....-...))))). ( -15.66, z-score = -0.82, R) >droSec1.super_0 8039698 94 - 21120651 -------------------------CUCUCUUAAAAUAAAAUCAUUAUUUUACAAAACAUUUCCAAGAUUCCUCAAAGCAAUUAUCCACUUGCUGUCUGGACUGCUUCGUUUUGCAGUA -------------------------......((((((((.....))))))))..................((....(((((........)))))....))(((((........))))). ( -15.50, z-score = -2.42, R) >droSim1.chr3L 15277096 79 - 22553184 -------------------------CUCUUUUAAAAUAAAAU---------------CAUUUCCAAGAUUCCUCAAAGCAAUUAUCCACUUGCUGUCUGGGUUGCUUCGUUUUGCAGUA -------------------------.................---------------............(((....(((((........)))))....)))((((........)))).. ( -9.90, z-score = -0.08, R) >consensus _________________________CUCUAUUAAAAUAAAAU_AUCGAUUUACAAAACAUUUCCAAGAUUCCUCAAAGCAAUUAUCCACUUGCUGUCUGGACUGCUUCG_UUUGCAGUA .....................................................................(((....(((((........)))))....)))((((........)))).. (-11.40 = -11.28 + -0.12)

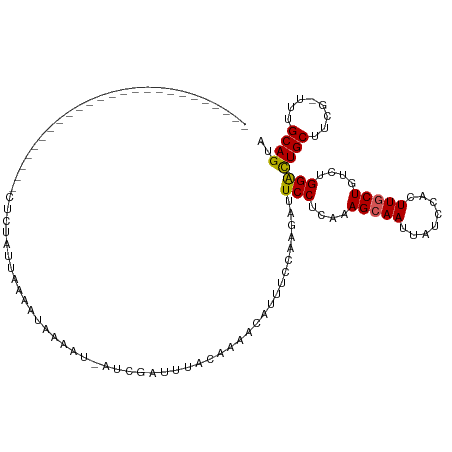
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:18 2011