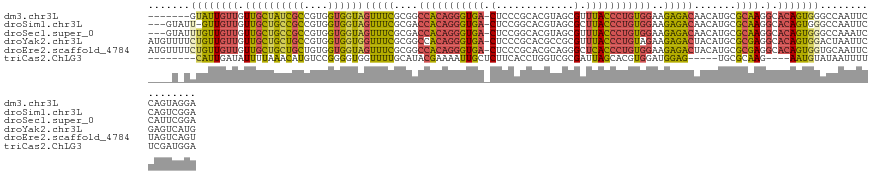
| Sequence ID | dm3.chr3L |
|---|---|
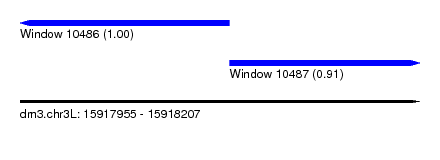
| Location | 15,917,955 – 15,918,207 |
| Length | 252 |
| Max. P | 0.995719 |

| Location | 15,917,955 – 15,918,087 |
|---|---|
| Length | 132 |
| Sequences | 5 |
| Columns | 133 |
| Reading direction | reverse |
| Mean pairwise identity | 97.74 |
| Shannon entropy | 0.03819 |
| G+C content | 0.49245 |
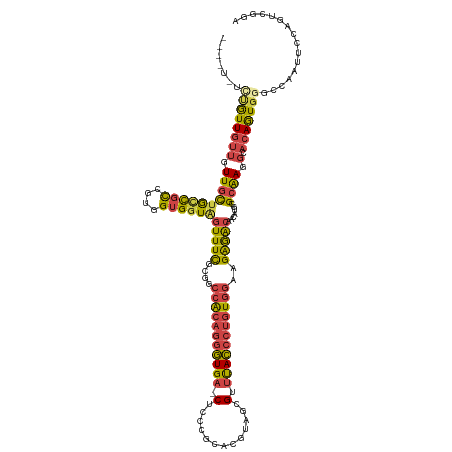
| Mean single sequence MFE | -43.04 |
| Consensus MFE | -41.12 |
| Energy contribution | -41.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.995719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15917955 132 - 24543557 GGCAAGUGCUUUCUCUCAGAGAGCGCUCUCUUCCUUUUGGCACGGCACGC-UUUUACAGUUAUACAGCAGAGCGCGGCUUUUCACAAGCCUCGCUCCAUAAAAUCAUUCAGUUUGUAGACAACGUUUGCCGUG ((..(((((((((.....)))))))))..)).((....))(((((((.((-.(((((((((....))).(((((.(((((.....))))).))))).................))))))....)).))))))) ( -43.40, z-score = -3.22, R) >droEre2.scaffold_4784 18171436 133 - 25762168 GGAACGUGCUUUCUCUCAGAGAGCGCUCUCUUCCUUUUGGCACGCCACGCUUUUUACAGUUAUACAGCAGAGCGCGGCUUUUCGCAAGCCUCGCUCCAUAAAAUCAUUCAGUUUGUAGACAACGUUUGCCGUG ((((.((((((((.....))))))))....))))....((((....(((...(((((((((....))).(((((.(((((.....))))).))))).................))))))...))).))))... ( -39.90, z-score = -2.05, R) >droYak2.chr3L 18520439 133 - 24197627 GGAACGUGCUUUCUCUCAGAGAGCGCUCUCUUCCUUUUGCCACGGCACGCUUUUUACAGUUAUACAGCAGAGCGCGGCUUUUCACAAGCCUCGCUCCAUAAAAUCAUUCAGUUUGUAGACAACGUUUGCCGUG ((((.((((((((.....))))))))....))))......(((((((.(((((((((((((....))).(((((.(((((.....))))).))))).................)))))).)).)).))))))) ( -45.10, z-score = -4.26, R) >droSec1.super_0 8024888 132 - 21120651 GGCAAGUGCUUUCUCUCAGAGAGCGCUCUCUUCCUUUUGGCACGGCACGC-UUUUACAGUUAUACAGCAGAGCGCGGCUUUUCACAAGCCUCGCUCCAUAAAAUCAUUCAGUUUGUAGACAACGUUUGCCGUG ((..(((((((((.....)))))))))..)).((....))(((((((.((-.(((((((((....))).(((((.(((((.....))))).))))).................))))))....)).))))))) ( -43.40, z-score = -3.22, R) >droSim1.chr3L 15264176 132 - 22553184 GGCAAGUGCUUUCUCUCAGAGAGCGCUCUCUUCCUUUUGGCACGGCACGC-UUUUACAGUUAUACAGCAGAGCGCGGCUUUUCACAAGCCUCGCUCCAUAAAAUCAUUCAGUUUGUAGACAACGUUUGCCGUG ((..(((((((((.....)))))))))..)).((....))(((((((.((-.(((((((((....))).(((((.(((((.....))))).))))).................))))))....)).))))))) ( -43.40, z-score = -3.22, R) >consensus GGCAAGUGCUUUCUCUCAGAGAGCGCUCUCUUCCUUUUGGCACGGCACGC_UUUUACAGUUAUACAGCAGAGCGCGGCUUUUCACAAGCCUCGCUCCAUAAAAUCAUUCAGUUUGUAGACAACGUUUGCCGUG .....((((((((.....))))))))..............(((((((.((..(((((((((....))).(((((.(((((.....))))).))))).................))))))....)).))))))) (-41.12 = -41.32 + 0.20)
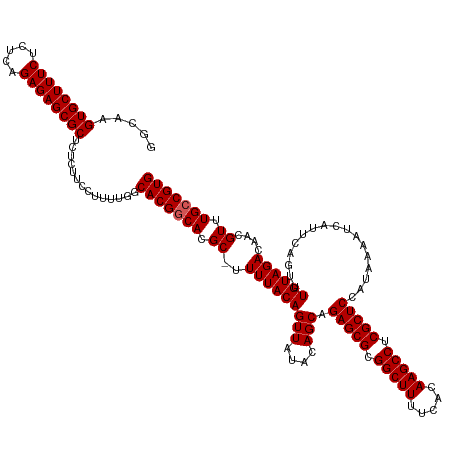
| Location | 15,918,087 – 15,918,207 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 71.93 |
| Shannon entropy | 0.53393 |
| G+C content | 0.53893 |
| Mean single sequence MFE | -45.62 |
| Consensus MFE | -27.07 |
| Energy contribution | -27.22 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.51 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15918087 120 + 24543557 -------GUAUUGUUGUUGCUAUCGCCGUGGUGGUAGUUUCGCGGCCACAGGGUGA-CUCCCGCACGUAGCGUUUACCCUGUGGAAGAGACAACAUGCGCAAGGCACAGUGGGCCAAUUCCAGUAGGA -------((((.((((((((((((((....)))))))).((....(((((((((((-....(((.....))).)))))))))))..))))))))))))....(((.......)))...(((....))) ( -47.10, z-score = -1.80, R) >droSim1.chr3L 15264308 123 + 22553184 ---GUAUU-GUUGUUGUUGCUGCCGCCGUGGUGGUAGUUUCGCGACCACAGGGUGA-CUCCGGCACGUAGCGCUUACCCUGUGGAAGAGACAACAUGCGCAAGGCACAGUGGGCCAAUUCCAGUCGGA ---((((.-(((((((..((((((((....))))))))..)....(((((((((((-.....((.....))..)))))))))))....))))))))))....(((.......)))...(((....))) ( -48.00, z-score = -1.01, R) >droSec1.super_0 8025020 124 + 21120651 ---GUAUUUGUUGUUGUUGCUGCCGCCGUGGUGGUAGUUUCGCGACCACAGGGUGA-CUCCGGCACGUAGCGUUUACCCUGUGGAAGAGACAACAUGCGCAAGGCACAGUGGGCCAAAUCCAUUCGGA ---((...((((((((..((((((((....))))))))..)....(((((((((((-.....((.....))..)))))))))))....)))))))...))..(((.......)))...(((....))) ( -47.10, z-score = -1.21, R) >droYak2.chr3L 18520572 127 + 24197627 AUGUUUUCUGUUGUUGUUGCUGCUGCCGUGGUGGUGGUUUCGCGGCCACAGGGUGA-CUCCCGCACGCCGCGUUUACCCUGUAGAAGAGACUACAUGCGCGAGGCACAGUGGACUAAUUCGAGUCAUG (((.(((..((((...(..(((.((((.(((((((((((((....(.(((((((((-....(((.....))).))))))))).)..)))))))).))).)).)))))))..)..))))..))).))). ( -43.70, z-score = -0.06, R) >droEre2.scaffold_4784 18171569 127 + 25762168 AUGUUUUCUGUUGUUGUUGCUGCUGCUGUGGUGGUAGUUUCGCGGCCACAGGGUGA-CUCCCGCACGCAGGGCUCACCCUGUGGAAGAGACUACAUGCGCGAGGCACAGUGGUGCAAUUCUAGUCAGU .......((((((..(((((.((..(((((...((((((((....(((((((((((-..(((.......))).)))))))))))..))))))))...(....).)))))..)))))))..))).))). ( -59.90, z-score = -3.54, R) >triCas2.ChLG3 7310182 111 - 32080666 --------CAUUGAUAUUUUAAACAUGUCCGGGGUGGUUUUGCAUACGAAAAUUGCUCUUCACCUGGUCGCGAUUAGCACGUGGAUGGAG-----UGCGCAAG----AAUGUAUAAUUUUUCGAUGGA --------(((((((((((((..(((((.(.((((((....(((.........)))...)))))).)..((.....)))))))..)))))-----))(....)----.............)))))).. ( -27.90, z-score = -0.30, R) >consensus ____U_UCUGUUGUUGUUGCUGCCGCCGUGGUGGUAGUUUCGCGGCCACAGGGUGA_CUCCCGCACGUAGCGUUUACCCUGUGGAAGAGACAACAUGCGCAAGGCACAGUGGGCCAAUUCCAGUCGGA .......((((((((.((((((((((....))))))(((((....(((((((((((.(.............).)))))))))))..))))).......)))).).)))))))................ (-27.07 = -27.22 + 0.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:17 2011