| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,898,025 – 15,898,150 |
| Length | 125 |
| Max. P | 0.999495 |

| Location | 15,898,025 – 15,898,150 |
|---|---|
| Length | 125 |
| Sequences | 5 |
| Columns | 125 |
| Reading direction | forward |
| Mean pairwise identity | 80.54 |
| Shannon entropy | 0.34242 |
| G+C content | 0.29032 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -24.62 |
| Energy contribution | -24.62 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.84 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.80 |
| SVM RNA-class probability | 0.999333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
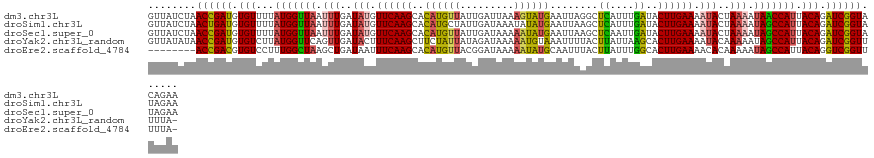
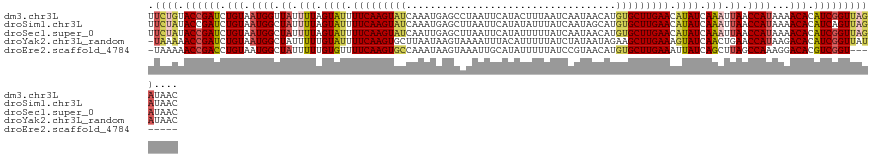
>dm3.chr3L 15898025 125 + 24543557 GUUAUCUAACCGAUGUGUUUUAUGGUUAAUUUGAUAUGUUCAAGCACAUGUUAUUGAUUAAAGUAUGAAUUAGGCUCAUUUGAUACUUGAAAAUACUAAAAUAACCAUUACAGAUCGGUACAGAA ....(((.((((((.(((...(((((((.((((.(((.((((((((.(((...((((((........))))))...))).))...)))))).))).)))).))))))).))).))))))..))). ( -33.60, z-score = -4.08, R) >droSim1.chr3L 15244083 125 + 22553184 GUUAUCUAACUGAUGUGUUUUAUGGUUAAUUUGAUAUGUUCAAGCACAUGCUAUUGAUAAAUAUAUGAAUUAAGCUCAUUUGAUACUUGAAAAUACUAAAAUAGCCAUUACAGAUCGGUAUAGAA ....((((((((((.(((...(((((((.((((.(((.((((((..((((.((((....)))))))).((((((....)))))).)))))).))).)))).))))))).))).)))))).)))). ( -32.80, z-score = -3.95, R) >droSec1.super_0 8005034 125 + 21120651 GUUAUCUAACCGAUGUGUUUUAUGGUUAAUUUGAUAUGUUCAAGCACAUGUUAUUGAUAAAAAUAUGAAUUAAGCUCAAUUGAUACUUGAAAAUACUAAAAUAGCCAUUACAGAUCGGUAUAGAA ....((((((((((.(((...(((((((.((((.(((.((((((..((((((.........)))))).(((((......))))).)))))).))).)))).))))))).))).)))))).)))). ( -35.30, z-score = -5.23, R) >droYak2.chr3L_random 3036267 124 - 4797643 GUUAUAUAACCGAUGUGUCUUAUGGUUCAGUUGAUACUUUCAAGCUUCUAUUAUAGAUAAAAAUGUAAAUUUUACUUAUUAAGCACUUGAAAAUACAAAAAUAGCCAUUACAGAUCGGUUUUUA- .......(((((((.(((...((((((...(((.((.(((((((...((...((((.(((((.......))))).))))..))..))))))).)))))....)))))).))).)))))))....- ( -26.80, z-score = -2.98, R) >droEre2.scaffold_4784 18151057 116 + 25762168 --------ACCGACGUGUCCUUUGGCUAAGCUGAUAAUUUCAAGCACAUGUUACGGAUAAAAAUAUGCAAUUUACUUAUUUGGCACUUGAAAACACAAAAAUAGCCAUUACAGGUCGGUUUUUA- --------((((((.(((....((((((.(((((.....)).))).((.((..(((((((((((.....))))..)))))))..)).))............))))))..))).)))))).....- ( -29.70, z-score = -2.97, R) >consensus GUUAUCUAACCGAUGUGUUUUAUGGUUAAUUUGAUAUGUUCAAGCACAUGUUAUUGAUAAAAAUAUGAAUUAAGCUCAUUUGAUACUUGAAAAUACUAAAAUAGCCAUUACAGAUCGGUAUAGAA ........((((((.(((...(((((((.(((..(((.((((((..((((((.........))))))........((....))..)))))).)))..))).))))))).))).))))))...... (-24.62 = -24.62 + -0.00)
| Location | 15,898,025 – 15,898,150 |
|---|---|
| Length | 125 |
| Sequences | 5 |
| Columns | 125 |
| Reading direction | reverse |
| Mean pairwise identity | 80.54 |
| Shannon entropy | 0.34242 |
| G+C content | 0.29032 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -20.51 |
| Energy contribution | -21.75 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.23 |
| Mean z-score | -4.09 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.94 |
| SVM RNA-class probability | 0.999495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
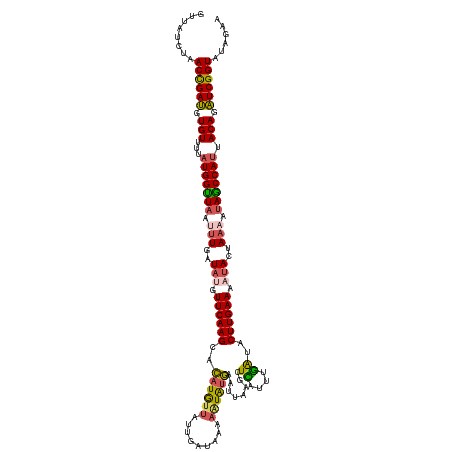
>dm3.chr3L 15898025 125 - 24543557 UUCUGUACCGAUCUGUAAUGGUUAUUUUAGUAUUUUCAAGUAUCAAAUGAGCCUAAUUCAUACUUUAAUCAAUAACAUGUGCUUGAACAUAUCAAAUUAACCAUAAAACACAUCGGUUAGAUAAC .((((.((((((.(((.(((((((.(((.((((.(((((((((....(((...(((........))).))).......))))))))).)))).))).)))))))...))).)))))))))).... ( -32.10, z-score = -5.28, R) >droSim1.chr3L 15244083 125 - 22553184 UUCUAUACCGAUCUGUAAUGGCUAUUUUAGUAUUUUCAAGUAUCAAAUGAGCUUAAUUCAUAUAUUUAUCAAUAGCAUGUGCUUGAACAUAUCAAAUUAACCAUAAAACACAUCAGUUAGAUAAC .((((.((.(((.(((.((((.((.(((.((((.((((((((((....))(((..(((.(((....))).))))))...)))))))).)))).))).)).))))...))).))).)))))).... ( -23.30, z-score = -2.39, R) >droSec1.super_0 8005034 125 - 21120651 UUCUAUACCGAUCUGUAAUGGCUAUUUUAGUAUUUUCAAGUAUCAAUUGAGCUUAAUUCAUAUUUUUAUCAAUAACAUGUGCUUGAACAUAUCAAAUUAACCAUAAAACACAUCGGUUAGAUAAC .((((.((((((.(((.((((.((.(((.((((.(((((((((..(((((....(((....)))....))))).....))))))))).)))).))).)).))))...))).)))))))))).... ( -30.80, z-score = -5.25, R) >droYak2.chr3L_random 3036267 124 + 4797643 -UAAAAACCGAUCUGUAAUGGCUAUUUUUGUAUUUUCAAGUGCUUAAUAAGUAAAAUUUACAUUUUUAUCUAUAAUAGAAGCUUGAAAGUAUCAACUGAACCAUAAGACACAUCGGUUAUAUAAC -....(((((((.(((.((((........((((((((((((.((..(((..(((((.......)))))..)))...))..))))))))))))........))))...))).)))))))....... ( -29.69, z-score = -4.31, R) >droEre2.scaffold_4784 18151057 116 - 25762168 -UAAAAACCGACCUGUAAUGGCUAUUUUUGUGUUUUCAAGUGCCAAAUAAGUAAAUUGCAUAUUUUUAUCCGUAACAUGUGCUUGAAAUUAUCAGCUUAGCCAAAGGACACGUCGGU-------- -.....((((((.(((..((((((..((.(((.(((((((..(....((.(((((.........)))))...))....)..))))))).))).))..))))))....))).))))))-------- ( -31.30, z-score = -3.24, R) >consensus UUCUAUACCGAUCUGUAAUGGCUAUUUUAGUAUUUUCAAGUAUCAAAUGAGCAUAAUUCAUAUUUUUAUCAAUAACAUGUGCUUGAACAUAUCAAAUUAACCAUAAAACACAUCGGUUAGAUAAC .((((.((((((.(((.((((.((.(((.((((.(((((((((...................................))))))))).)))).))).)).))))...))).)))))))))).... (-20.51 = -21.75 + 1.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:16 2011