
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,883,736 – 15,883,844 |
| Length | 108 |
| Max. P | 0.936789 |

| Location | 15,883,736 – 15,883,844 |
|---|---|
| Length | 108 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.94 |
| Shannon entropy | 0.18212 |
| G+C content | 0.48047 |
| Mean single sequence MFE | -33.31 |
| Consensus MFE | -23.85 |
| Energy contribution | -24.64 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
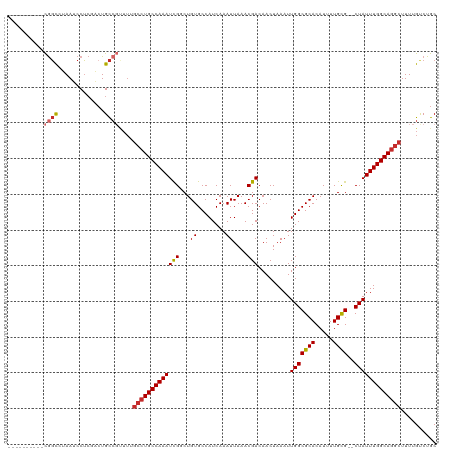
>dm3.chr3L 15883736 108 + 24543557 ----------CGGCCUAAUCCUCCACUGCCGUUUUGCCUGCCAAAUCGGCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGCG--CUAUUUGGCAGGCUUUUGCCUGC ----------((((.............))))....((((((((((..(((...))).......................(((((((.....))))--))))))))))))).......... ( -34.52, z-score = -2.28, R) >droEre2.scaffold_4784 18137534 108 + 25762168 ----------CGGCCUAAUCCUCUACUGCCGUUUUGCCUGCCAAAUCGGCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGCG--CUAUUUGGCAGGCUUUUGUCUGC ----------((((.............))))....((((((((((..(((...))).......................(((((((.....))))--))))))))))))).......... ( -34.52, z-score = -2.91, R) >droYak2.chr3L 18495420 108 + 24197627 ----------CGGCCUAAUCCUCCACUGCCGUUUUGCCUGCCAAAUCGGCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGCG--CUAUUUGGCAGGCUUUUGUCUGC ----------((((.............))))....((((((((((..(((...))).......................(((((((.....))))--))))))))))))).......... ( -34.52, z-score = -2.62, R) >droSec1.super_0 7991304 100 + 21120651 ----------CGGCCUAAUCCUCCACUGC--------CUGCCAAAUCGGCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGCG--CUAUUUGGCAGGCUUUUGUCUGC ----------.................((--------((((((((..(((...))).......................(((((((.....))))--))))))))))))).......... ( -30.80, z-score = -2.43, R) >droSim1.chr3L 15229473 108 + 22553184 ----------CGGCCUAAUCCUCUACUGCCGUUUUGCCUGCCAAAUCGGCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGCG--CUAUUUGGCAGGCUUUUGUCUGC ----------((((.............))))....((((((((((..(((...))).......................(((((((.....))))--))))))))))))).......... ( -34.52, z-score = -2.91, R) >dp4.chrXR_group6 3645918 108 + 13314419 ----------CGGCGUAAUCCUCGACUGCCGCUUUGCCUGCCAAAUCGUCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGUG--CUAUUUGGCAGGCUUUUGUCUGC ----------((((((........)).))))....(((((((((((.......((((......................))))((((.....)))--).))))))))))).......... ( -33.95, z-score = -2.66, R) >droPer1.super_9 1941758 108 + 3637205 ----------CGGCGUAAUCCUCGACUGCCGCUUUGCCUGCCAAAUCGUCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGUG--CUAUUUGGCAGGCUUUUGUCUGC ----------((((((........)).))))....(((((((((((.......((((......................))))((((.....)))--).))))))))))).......... ( -33.95, z-score = -2.66, R) >droAna3.scaffold_13337 12026060 108 - 23293914 ----------CAGCGUAAUCCUAGGGUGCCGUUUUGCCUGCCAAAUCGGCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGCG--CUAUUUGGCAAGCUUUUGUCUGC ----------.((((..(((....)))..))))..((.(((((((..(((...))).......................(((((((.....))))--)))))))))).)).......... ( -29.00, z-score = -0.04, R) >droWil1.scaffold_180727 1352327 119 - 2741493 CCCUCUUUGUUGUUCUAAUCCGCGCCUGUC-UUUUGCCUGCCAAAUUGUCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGUGUGCUAUUUGGCAGGCUUUUGUCUGC .......................((..(.(-....(((((((((((((((.((.............)))))))......((((((((.....))))))))))))))))))....).).)) ( -34.02, z-score = -3.50, R) >consensus __________CGGCCUAAUCCUCGACUGCCGUUUUGCCUGCCAAAUCGGCUGUGCCAUCAAAUUAAACGACAACUAAAUUGGCGCACAUUUUGCG__CUAUUUGGCAGGCUUUUGUCUGC ..........((((.............))))....(((((((((((.(..(((((((......................)))))))(.....)....).))))))))))).......... (-23.85 = -24.64 + 0.79)

| Location | 15,883,736 – 15,883,844 |
|---|---|
| Length | 108 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.94 |
| Shannon entropy | 0.18212 |
| G+C content | 0.48047 |
| Mean single sequence MFE | -36.59 |
| Consensus MFE | -28.51 |
| Energy contribution | -28.89 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
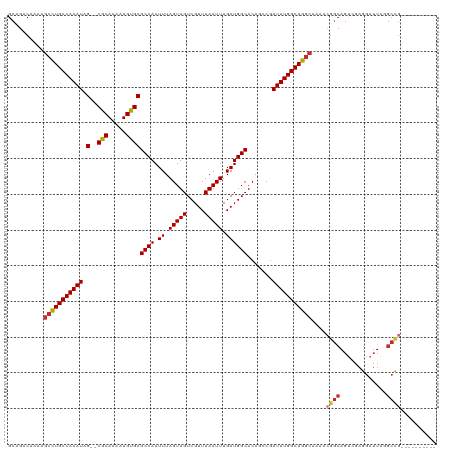
>dm3.chr3L 15883736 108 - 24543557 GCAGGCAAAAGCCUGCCAAAUAG--CGCAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGCCGAUUUGGCAGGCAAAACGGCAGUGGAGGAUUAGGCCG---------- ...(((....(((((((((((.(--((((.....))))).......((..(((.......)))..)).......)))))))))))...((....))..........))).---------- ( -38.40, z-score = -1.83, R) >droEre2.scaffold_4784 18137534 108 - 25762168 GCAGACAAAAGCCUGCCAAAUAG--CGCAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGCCGAUUUGGCAGGCAAAACGGCAGUAGAGGAUUAGGCCG---------- ..........(((((((((((.(--((((.....))))).......((..(((.......)))..)).......)))))))))))....((((.............))))---------- ( -37.62, z-score = -2.71, R) >droYak2.chr3L 18495420 108 - 24197627 GCAGACAAAAGCCUGCCAAAUAG--CGCAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGCCGAUUUGGCAGGCAAAACGGCAGUGGAGGAUUAGGCCG---------- ..........(((((((((((.(--((((.....))))).......((..(((.......)))..)).......)))))))))))....((((.............))))---------- ( -37.62, z-score = -2.18, R) >droSec1.super_0 7991304 100 - 21120651 GCAGACAAAAGCCUGCCAAAUAG--CGCAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGCCGAUUUGGCAG--------GCAGUGGAGGAUUAGGCCG---------- .....((...(((((((((((.(--((((.....))))).......((..(((.......)))..)).......)))))))))--------))..))..((......)).---------- ( -34.60, z-score = -2.11, R) >droSim1.chr3L 15229473 108 - 22553184 GCAGACAAAAGCCUGCCAAAUAG--CGCAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGCCGAUUUGGCAGGCAAAACGGCAGUAGAGGAUUAGGCCG---------- ..........(((((((((((.(--((((.....))))).......((..(((.......)))..)).......)))))))))))....((((.............))))---------- ( -37.62, z-score = -2.71, R) >dp4.chrXR_group6 3645918 108 - 13314419 GCAGACAAAAGCCUGCCAAAUAG--CACAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGACGAUUUGGCAGGCAAAGCGGCAGUCGAGGAUUACGCCG---------- ..........(((((((((((.(--(((.....))))((((.((.(((((.....))))).)))))).......)))))))))))....((((((((...))))..))))---------- ( -38.10, z-score = -3.24, R) >droPer1.super_9 1941758 108 - 3637205 GCAGACAAAAGCCUGCCAAAUAG--CACAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGACGAUUUGGCAGGCAAAGCGGCAGUCGAGGAUUACGCCG---------- ..........(((((((((((.(--(((.....))))((((.((.(((((.....))))).)))))).......)))))))))))....((((((((...))))..))))---------- ( -38.10, z-score = -3.24, R) >droAna3.scaffold_13337 12026060 108 + 23293914 GCAGACAAAAGCUUGCCAAAUAG--CGCAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGCCGAUUUGGCAGGCAAAACGGCACCCUAGGAUUACGCUG---------- ..........(((((((((((.(--((((.....))))).......((..(((.......)))..)).......)))))))))))....((((.............))))---------- ( -32.82, z-score = -1.15, R) >droWil1.scaffold_180727 1352327 119 + 2741493 GCAGACAAAAGCCUGCCAAAUAGCACACAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGACAAUUUGGCAGGCAAAA-GACAGGCGCGGAUUAGAACAACAAAGAGGG ((........(((((((((((.(((((.....)))))((((.((.(((((.....))))).)))))).......)))))))))))....-.....))....................... ( -34.43, z-score = -2.91, R) >consensus GCAGACAAAAGCCUGCCAAAUAG__CGCAAAAUGUGCGCCAAUUUAGUUGUCGUUUAAUUUGAUGGCACAGCCGAUUUGGCAGGCAAAACGGCAGUCGAGGAUUAGGCCG__________ ..........(((((((((((....(((.....))).((((.((.(((((.....))))).)))))).......)))))))))))....((((.............)))).......... (-28.51 = -28.89 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:14 2011