| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,866,232 – 15,866,324 |
| Length | 92 |
| Max. P | 0.923265 |

| Location | 15,866,232 – 15,866,324 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.87 |
| Shannon entropy | 0.51276 |
| G+C content | 0.56396 |
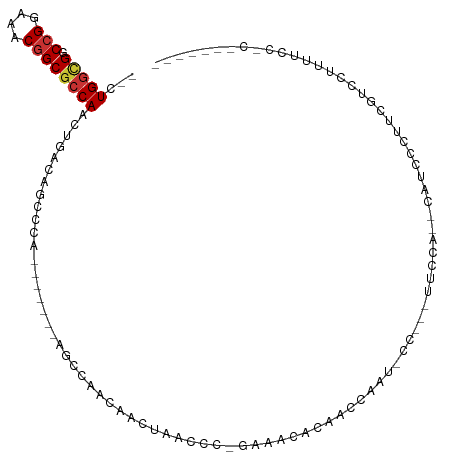
| Mean single sequence MFE | -19.20 |
| Consensus MFE | -14.34 |
| Energy contribution | -14.62 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
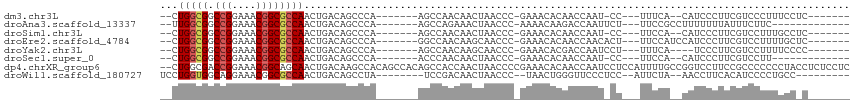
>dm3.chr3L 15866232 92 + 24543557 --CUGGCGGCCGGAAACGGCGCCAACUGACAGCCCA-------AGCCAACAACUAACCC-GAAACACAACCAAU-CC---UUUCA--CAUCCCUUCGUCCCUUUCCUC------- --.(((((.(((....))))))))...(((.((...-------.)).............-((((..........-..---)))).--.........))).........------- ( -19.00, z-score = -1.86, R) >droAna3.scaffold_13337 12009833 89 - 23293914 --UUGGCGGCCGGAAACGGCGCCAACUGACAGCCCA-------AGCCAGAAACUAACCC-AAAACAAGACCAAUUCU---UUCCGCCUUUUUUUAUUUCUUC------------- --..((((((((....)))).....(((...((...-------.)))))..........-.................---...))))...............------------- ( -19.30, z-score = -1.28, R) >droSim1.chr3L 15211465 92 + 22553184 --CUGGCGGCCGGAAACGGCGCCAACUGACAGCCCA-------AGCCAACAACUAACCC-GAAACACAACCAAU-CC---UUCCA--CAUCCCUUCGUCCUUUGCCUC------- --.(((((.(((....))))))))...(((.((...-------.)).............-(((...........-..---.....--......)))))).........------- ( -17.66, z-score = -0.76, R) >droEre2.scaffold_4784 18119372 95 + 25762168 --CUGGCGGCCGGAAACGGCGCCAACUGACAGCCCA-------GGCCAACAAGCAACCC-GAAACACAACCAACACU---UUCCAUCCAUCCCUUCGUCCUUUUGCUC------- --.((((.((((....)))).....(((......))-------)))))...(((((..(-(((..............---.............)))).....))))).------- ( -21.63, z-score = -0.81, R) >droYak2.chr3L 18477881 91 + 24197627 --CUGGCGGCCGGAAACGGCGCCAACUGACAGCCCA-------AGCCAACAAGCAACCC-GAAACACGACCAAUCCU---UUUCA----UCCCUUCGUCCUUUUCCCC------- --.(((((.(((....))))))))...(((......-------.((......)).....-((((...((....))..---)))).----.......))).........------- ( -20.50, z-score = -1.25, R) >droSec1.super_0 7973687 86 + 21120651 --CUGGCGGCCGGAAACGGCGCCAACUGACAGCCCA-------ACCCAACAACUAACCC-GAAACACAACCAAU-CC---UUCCA--CAUCCCUUCGUCCUU------------- --.(((((.(((....))))))))............-------................-..............-..---.....--...............------------- ( -16.70, z-score = -1.55, R) >dp4.chrXR_group6 3629461 113 + 13314419 --CUGGCGACCGGAAACGGCAGCAACUGACAAGCCACAGCCACAGCCACCAACUAACCCCGAAACACAACCAAUCCUCCAUUUUGCCGGUCCUUCCGCCCCCCCUACCUCUCCUC --..(((((((((....(((.....(((........))).....))).......................(((.........))))))))).....)))................ ( -18.10, z-score = -0.05, R) >droWil1.scaffold_180727 1410238 92 + 2741493 UCCUGGUGGCAGGAAACGGCGCCAACUGACAGCCUA--------UCCGACAACUAACCC--UAACUGGGUUCCCUCC--AUUCUA--AACCUUCACAUCCCCUGCC--------- .......((((((....(((...........)))..--------..........(((((--.....)))))......--......--.............))))))--------- ( -20.70, z-score = -0.82, R) >consensus __CUGGCGGCCGGAAACGGCGCCAACUGACAGCCCA_______AGCCAACAACUAACCC_GAAACACAACCAAU_CC___UUCCA__CAUCCCUUCGUCCUUUUCC_C_______ ...(((((.(((....))))))))........................................................................................... (-14.34 = -14.62 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:09 2011