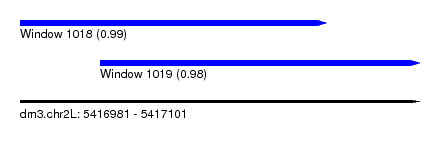
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,416,981 – 5,417,101 |
| Length | 120 |
| Max. P | 0.991981 |

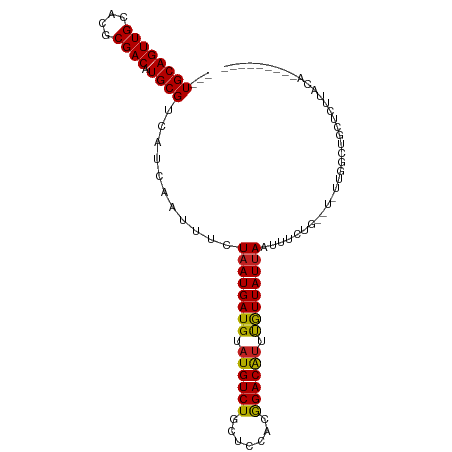
| Location | 5,416,981 – 5,417,073 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.55 |
| Shannon entropy | 0.43114 |
| G+C content | 0.41536 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -17.68 |
| Energy contribution | -17.47 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.991981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5416981 92 + 23011544 ---UGCAGUUGCACGCGACAUGCGUCAUCAAUUUCUAAUGAUGUAUGUCUGCUCCACGGACAUUUGUUAUUAAUUUCUG--U-UGGGCUGCUCUUACA--------- ---.(((((.....((((((((((((((.........)))))))))))).)).(((((((.(((.......))).))))--.-)))))))).......--------- ( -30.00, z-score = -3.50, R) >droSim1.chr2L 5209566 92 + 22036055 ---UGCAGUUGCACGCGACAUGCGUCAUCAAUUUCUAAUGAUGUAUGUCUGCUCCACGGACAUUUGUUAUUAAUUUCUG--U-UGGGCUGCUCUUACA--------- ---.(((((.....((((((((((((((.........)))))))))))).)).(((((((.(((.......))).))))--.-)))))))).......--------- ( -30.00, z-score = -3.50, R) >droSec1.super_5 3491667 92 + 5866729 ---UGCAGUUGCACGCGACAUGCGUCAUCAAUUUCUAAUGAUGUAUGUCUGCUCCACGGACAUUUGUUAUUAAUUUCUG--U-UGGGCUGCUCUUACA--------- ---.(((((.....((((((((((((((.........)))))))))))).)).(((((((.(((.......))).))))--.-)))))))).......--------- ( -30.00, z-score = -3.50, R) >droYak2.chr2L 8543364 92 - 22324452 ---UGCAGUUGCACGCGACAUGCGUCAUCAAUUUCUAAUGAUGUAUGUCUGCUCCACGGACAUUUGUUAUUAAUUUCUG--U-UGGCCCGCUCUUACA--------- ---(((....))).((((((((((((((.........)))))))))))).(((..(((((.(((.......))).))))--)-.)))..)).......--------- ( -24.90, z-score = -2.03, R) >droEre2.scaffold_4929 5499049 92 + 26641161 ---UGCAGUUGCACGCGACAUGCGUCAUCAAUUUCUAAUGAUGUAUGUCUGCUCCACGGACAUUUGUUAUUAAUUUCUG--U-UGGGCUGCUCUUACA--------- ---.(((((.....((((((((((((((.........)))))))))))).)).(((((((.(((.......))).))))--.-)))))))).......--------- ( -30.00, z-score = -3.50, R) >droAna3.scaffold_12916 6442357 94 - 16180835 ---UGCAGUUGCACGCGACAUGCGUCAUCAAUUUCUAAUGAUGUAUGUCUGCUCCACGGACGUUUGUUAUUAAUUUUCAGUU-CUGGCCGCUCUUACA--------- ---....((.((..((((((((((((((.........)))))))))))).)).(((.((((..................)))-))))..))....)).--------- ( -23.57, z-score = -1.21, R) >dp4.chr4_group1 4715473 106 + 5278887 AGUUGCAGUUGCACGCGACAUGCGUCAUCAAUUUCUAAUGAUGUAUGUCUGUGGAGCAGACAUUUGUUAUUAAUUCCAUUACACUGCCACUUCCACUGCUCUUUCU- (((.((((((((.(((((((((((((((.........)))))))))))).)))..)))(((....)))..............))))).)))...............- ( -29.30, z-score = -2.53, R) >droPer1.super_5 6311217 106 - 6813705 AGUUGCAGUUGCACGCGACAUGCGUCAUCAAUUUCUAAUGAUGUAUGUCUGUGGAGCAGACAUUUGUUAUUAAUUCCAUUACACUGCCACUUCCACUGCUCUUUCU- (((.((((((((.(((((((((((((((.........)))))))))))).)))..)))(((....)))..............))))).)))...............- ( -29.30, z-score = -2.53, R) >droVir3.scaffold_12963 342668 95 - 20206255 ---UGCAGUUGCACGCGACAUGCGUCAUCAAUUUCUAAUGAUGUAUGUCUUCAGCACGGACAUUUGUUAUGAAUUGUUAAUUGUUG-UAGCUUUUGUGC-------- ---.((((..((....((((((((((((.........))))))))))))..(((((..((((.((......)).))))...)))))-..))..))))..-------- ( -27.20, z-score = -1.35, R) >droMoj3.scaffold_6500 23327635 103 + 32352404 ---UGCAGUUGCACGCGACAUGCGUCAUCAAUUUCUAAUGAUGUCUGUCUUCAACACGGACAUUUGUUAUUAAUUGUUAAUUGUUG-UUGUACUGUUGCUGGUGUGC ---..((((.(((.((((((.(((....(((((..(((..((((((((.......))))))).)..)))..))))).....)))))-))))..))).))))...... ( -27.40, z-score = -0.87, R) >droGri2.scaffold_15252 5860459 104 - 17193109 ---UGCAGUUGCACGCGACAUGCGUCAUCAAUUUCUAAUGAUGUAUGUCUUCAGCAUGGACAUUCAUUAUUAAUUGUUAAUUGCUGCUUUUGCUGUUACUGUUGUGC ---.(((((.(((.((((((((((((((.........))))))))))))..(((((..((((............))))...))))).....))))).)))))..... ( -33.70, z-score = -3.09, R) >consensus ___UGCAGUUGCACGCGACAUGCGUCAUCAAUUUCUAAUGAUGUAUGUCUGCUCCACGGACAUUUGUUAUUAAUUUCUG__U_UUGGCUGCUCUUACA_________ ....(((((((....)))).)))............((((((((.((((((.......)))))).))))))))................................... (-17.68 = -17.47 + -0.21)

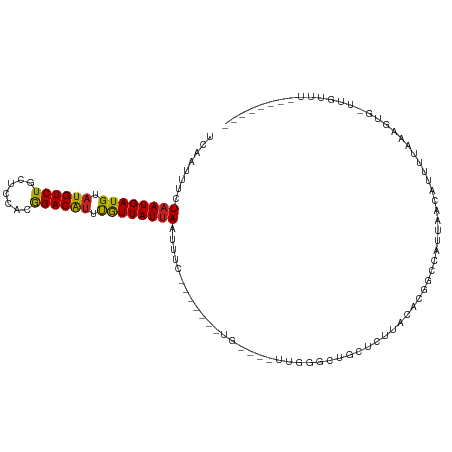
| Location | 5,417,005 – 5,417,101 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 72.11 |
| Shannon entropy | 0.56051 |
| G+C content | 0.33626 |
| Mean single sequence MFE | -23.21 |
| Consensus MFE | -10.48 |
| Energy contribution | -10.27 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5417005 96 + 23011544 UCAAUUUCUAAUGAUGUAUGUCUGCUCCACGGACAUUUGUUAUUAAUUUC-------UG----UUGGGCUGCUCUUACACGGCCAUUAACAUUUUAAAGUG-UUGUUU-------- .((((...((((((((.(((((((.....))))))).)))))))).....-------.)----)))(((((........)))))..(((((((....))))-)))...-------- ( -23.60, z-score = -2.76, R) >droSim1.chr2L 5209590 96 + 22036055 UCAAUUUCUAAUGAUGUAUGUCUGCUCCACGGACAUUUGUUAUUAAUUUC-------UG----UUGGGCUGCUCUUACACGGCCAUUAACAUUUUAAAGUG-UUGUUU-------- .((((...((((((((.(((((((.....))))))).)))))))).....-------.)----)))(((((........)))))..(((((((....))))-)))...-------- ( -23.60, z-score = -2.76, R) >droSec1.super_5 3491691 96 + 5866729 UCAAUUUCUAAUGAUGUAUGUCUGCUCCACGGACAUUUGUUAUUAAUUUC-------UG----UUGGGCUGCUCUUACACGGCCAUUAACAUUUUAAAGUG-UUGUUU-------- .((((...((((((((.(((((((.....))))))).)))))))).....-------.)----)))(((((........)))))..(((((((....))))-)))...-------- ( -23.60, z-score = -2.76, R) >droYak2.chr2L 8543388 96 - 22324452 UCAAUUUCUAAUGAUGUAUGUCUGCUCCACGGACAUUUGUUAUUAAUUUC-------UG----UUGGCCCGCUCUUACACGGCCAUUAACAUUUUAAAGUG-UUGUUU-------- ........((((((((.(((((((.....))))))).)))))))).....-------..----.(((((...........))))).(((((((....))))-)))...-------- ( -22.10, z-score = -2.48, R) >droEre2.scaffold_4929 5499073 96 + 26641161 UCAAUUUCUAAUGAUGUAUGUCUGCUCCACGGACAUUUGUUAUUAAUUUC-------UG----UUGGGCUGCUCUUACACGGCCAUUAACAUUUUAAAGUG-UUGUUU-------- .((((...((((((((.(((((((.....))))))).)))))))).....-------.)----)))(((((........)))))..(((((((....))))-)))...-------- ( -23.60, z-score = -2.76, R) >droAna3.scaffold_12916 6442381 98 - 16180835 UCAAUUUCUAAUGAUGUAUGUCUGCUCCACGGACGUUUGUUAUUAAUUUU-------CA--GUUCUGGCCGCUCUUACACGGCCAUUAACAUUUUAAAGAG-UUGUUU-------- .((((((.((((((((.(((((((.....))))))).)))))))).....-------..--(((.((((((........))))))..)))........)))-)))...-------- ( -25.40, z-score = -3.32, R) >dp4.chr4_group1 4715500 108 + 5278887 UCAAUUUCUAAUGAUGUAUGUCUGUGGAGCAGACAUUUGUUAUUAAUUCCAUUACACUGCCACUUCCACUGCUCUUUCUUAGCAAUUAACAAUUUAAAGUGGCUGUUU-------- ........((((((((.(((((((.....))))))).))))))))..........((.((((((.....((((.......)))).((((....)))))))))).))..-------- ( -24.50, z-score = -2.23, R) >droPer1.super_5 6311244 108 - 6813705 UCAAUUUCUAAUGAUGUAUGUCUGUGGAGCAGACAUUUGUUAUUAAUUCCAUUACACUGCCACUUCCACUGCUCUUUCUUAGCAAUUAACAAUUUAAAGUGGCUGUUU-------- ........((((((((.(((((((.....))))))).))))))))..........((.((((((.....((((.......)))).((((....)))))))))).))..-------- ( -24.50, z-score = -2.23, R) >droVir3.scaffold_12963 342692 107 - 20206255 UCAAUUUCUAAUGAUGUAUGUCUUCAGCACGGACAUUUGUUAUGAAUUGUUAAUUGUUG-UAGCUUU--------UGUGCAGACAUUAGUUAUUAAUAUUUUUUAAUGCACUUUUU ........(((((((..((((((...(((((((.....((((..(..(....)...)..-)))).))--------)))))))))))..)))))))..................... ( -18.60, z-score = -0.29, R) >droMoj3.scaffold_6500 23327659 115 + 32352404 UCAAUUUCUAAUGAUGUCUGUCUUCAACACGGACAUUUGUUAUUAAUUGUUAAUUGUUG-UUGUACUGUUGCUGGUGUGCGGACAUUAGUUAUUAAUAUGUUUUAAUGCACUUUUU ...........((((((((((...(((((..((((.(((....))).))))...)))))-...((((......)))).))))))))))((((((((......)))))).))..... ( -22.00, z-score = -0.69, R) >droGri2.scaffold_15252 5860483 107 - 17193109 UCAAUUUCUAAUGAUGUAUGUCUUCAGCAUGGACAUUCAUUAUUAAUUGUUAAUUGCUGCUUUUGCUGUUACUGUUGUGCGGACAUUAGUUAUUAAUAUUGUUUAAU--------- ........((((((((.((((((.......)))))).))))))))..(((((((....(((..((((((.((....)))))).))..))).))))))).........--------- ( -23.80, z-score = -1.90, R) >consensus UCAAUUUCUAAUGAUGUAUGUCUGCUCCACGGACAUUUGUUAUUAAUUUC_______UG____UUGGGCUGCUCUUACACGGCCAUUAACAUUUUAAAGUG_UUGUUU________ ........((((((((.((((((.......)))))).))))))))....................................................................... (-10.48 = -10.27 + -0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:52 2011