
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,849,333 – 15,849,463 |
| Length | 130 |
| Max. P | 0.757771 |

| Location | 15,849,333 – 15,849,430 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 86.46 |
| Shannon entropy | 0.25835 |
| G+C content | 0.69969 |
| Mean single sequence MFE | -43.10 |
| Consensus MFE | -35.01 |
| Energy contribution | -35.07 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.733320 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
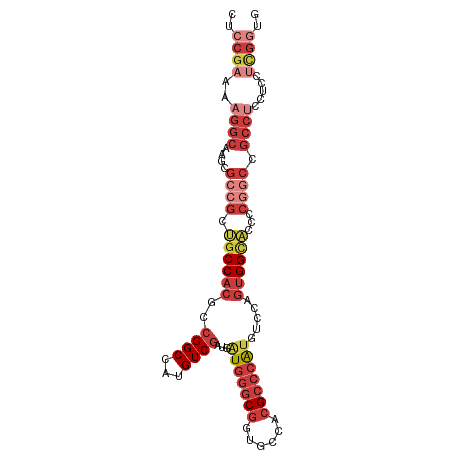
>dm3.chr3L 15849333 97 - 24543557 CAUGUCGAUGAUGGGCGGGGCCACGCCCAUGUCCAGUGGUACCCCGGCCGCCUCAUCCUCGGUGGCCACGCCCGGUACCCCAAGGAUCACCGAUCCG ...((((.((((((((((((((((...........))))..))))(((((((........)))))))..))))((....))....)))).))))... ( -43.60, z-score = -1.38, R) >droSim1.chr3L 15194249 97 - 22553184 CAUGUCGAUGAUGGGCGGUGCCACGCCCAUGUCCAGUGGCACCCCGGCCGCCUCCUCCUCGGUGGCCACGCCCGGCACCCCAAGGACCACCGAUCCG ...((((.....((((((((((((...........))))))))..(((((((........)))))))..))))((..((....)).))..))))... ( -44.10, z-score = -1.89, R) >droSec1.super_0 7957062 97 - 21120651 CAUGUCGAUGAUGGGCGGUGCCACGCCCAUGUCCAGUGGCACCCCGGCCGCCUCCUCCUCGGUGGCCACGCCCGGCACCCCAAGGACCACCGAUCCG ...((((.....((((((((((((...........))))))))..(((((((........)))))))..))))((..((....)).))..))))... ( -44.10, z-score = -1.89, R) >droYak2.chr3L 18461061 97 - 24197627 UAUGUCGAUGAUGGGCGGUGCCACGCCCAUGUCCAGUGGCACCCCGGCCGCCUCCUCCUCGGUGGCGACACCUGGAACCCCAAGGACCACCGAUCCA ...((((.(((((((((......)))))))((((.(((((......)))))....(((..((((....)))).))).......)))))).))))... ( -40.90, z-score = -1.61, R) >droEre2.scaffold_4784 18102546 97 - 25762168 UAUGUCGAUGAUGGGCGGUGCCACGCCCAUGUCCAGUGGCACCCCGGCCGCCUCCUCCUCGGUGGCCACGCCUGGAACCCCAAGGACCACCGAUCCG ...((((.(((((((((......)))))))((((...(((.....(((((((........)))))))..)))(((....))).)))))).))))... ( -41.90, z-score = -1.89, R) >droAna3.scaffold_13337 11985199 91 + 23293914 ---AAUGUCGGUGGGCGGGGUCACGCCCAUGUCAGGGGGUGCCACCCCGGCCACA---UCGGUGGCCACACCCGGAACCCCAAGGACCGUCGAUGCG ---..((.(((((((.((((((((.(((......))).)))..)))))((((((.---...))))))...)))((....))....)))).))..... ( -44.00, z-score = -0.44, R) >consensus CAUGUCGAUGAUGGGCGGUGCCACGCCCAUGUCCAGUGGCACCCCGGCCGCCUCCUCCUCGGUGGCCACGCCCGGAACCCCAAGGACCACCGAUCCG ...((((.(((((((((......)))))))((((...(((.....(((((((........)))))))..))).((....))..)))))).))))... (-35.01 = -35.07 + 0.06)



| Location | 15,849,366 – 15,849,463 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 78.91 |
| Shannon entropy | 0.43831 |
| G+C content | 0.68921 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -24.25 |
| Energy contribution | -25.89 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15849366 97 - 24543557 CUCCGAAAAGGCAAGCGCCGCUGCCACGCCGGCCAUGUCGAUGAUGGGCGGGGCCACGCCCAUGUCCAGUGGUACCCCGGCCGCCUCAUCCUCGGUG ..((((...((((.(.(((((......).))))).))))((((((((.(((((((((...........))))..))))).)))..))))).)))).. ( -40.50, z-score = -0.27, R) >droSim1.chr3L 15194282 97 - 22553184 CUCCGAAAAGGCAAGCGCCGCUGCCACGCCGGCCAUGUCGAUGAUGGGCGGUGCCACGCCCAUGUCCAGUGGCACCCCGGCCGCCUCCUCCUCGGUG ..((((..(((...((((((.((((((..((((...))))...(((((((......))))))).....))))))...))).)))..)))..)))).. ( -43.10, z-score = -1.57, R) >droSec1.super_0 7957095 97 - 21120651 CUCCGAAAAGGCAAGCGCCGCUGCCACGCCGGCCAUGUCGAUGAUGGGCGGUGCCACGCCCAUGUCCAGUGGCACCCCGGCCGCCUCCUCCUCGGUG ..((((..(((...((((((.((((((..((((...))))...(((((((......))))))).....))))))...))).)))..)))..)))).. ( -43.10, z-score = -1.57, R) >droYak2.chr3L 18461094 97 - 24197627 CUCCGAAAAGGCAAGCGCCGCUGCCACGCCGGCUAUGUCGAUGAUGGGCGGUGCCACGCCCAUGUCCAGUGGCACCCCGGCCGCCUCCUCCUCGGUG ..((((..(((...((((((.((((((..((((...))))...(((((((......))))))).....))))))...))).)))..)))..)))).. ( -43.10, z-score = -1.72, R) >droEre2.scaffold_4784 18102579 97 - 25762168 CUCCGAAAAGGCAAGCGCCGCUGCCACGCCGGCUAUGUCGAUGAUGGGCGGUGCCACGCCCAUGUCCAGUGGCACCCCGGCCGCCUCCUCCUCGGUG ..((((..(((...((((((.((((((..((((...))))...(((((((......))))))).....))))))...))).)))..)))..)))).. ( -43.10, z-score = -1.72, R) >droAna3.scaffold_13337 11985232 91 + 23293914 CUCCGAAAAGGCAAGCGCCACGGCCACGCCGGCAAUGUCG---GUGGGCGGGGUCACGCCCAUGUCAGGGGGUGCCACCCCGGCCACAUCGGUG--- ..((((...(((....)))..((((.(((((((...))))---)))...((((((((.(((......))).)))..)))))))))...))))..--- ( -47.90, z-score = -2.02, R) >droWil1.scaffold_180727 1307942 77 + 2741493 GUUCGAAAAGGCAAGCGCCGGAGCCACACCGGCCAUGUCAACAAUGACCCA---UACGCCAGCAACAGCUGGAUCGGCAA----------------- ((.(((...((((.(.(((((.......)))))).))))............---....(((((....))))).)))))..----------------- ( -24.80, z-score = -1.69, R) >consensus CUCCGAAAAGGCAAGCGCCGCUGCCACGCCGGCCAUGUCGAUGAUGGGCGGUGCCACGCCCAUGUCCAGUGGCACCCCGGCCGCCUCCUCCUCGGUG ..(((....(((....)))..((((((..((((...))))...(((((((......))))))).....))))))...))).((((........)))) (-24.25 = -25.89 + 1.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:08 2011