
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,840,219 – 15,840,335 |
| Length | 116 |
| Max. P | 0.535429 |

| Location | 15,840,219 – 15,840,335 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.81 |
| Shannon entropy | 0.53810 |
| G+C content | 0.44864 |
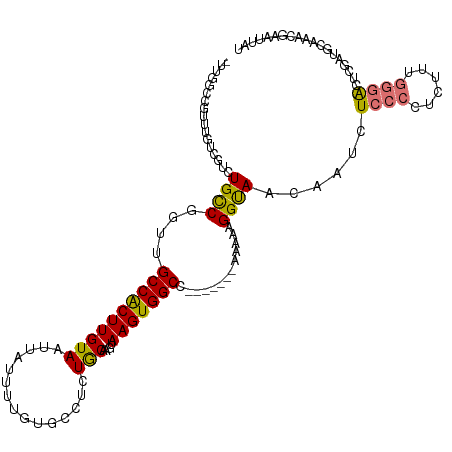
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -10.94 |
| Energy contribution | -11.04 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.535429 |
| Prediction | RNA |
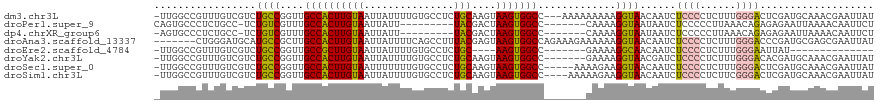
Download alignment: ClustalW | MAF
>dm3.chr3L 15840219 116 - 24543557 -UUGGCCGUUUGUCGUCUGCCGGUUGCCACUUGUAAUUAUUUUGUGCCUCUGCAAGUAAGUGGCC---AAAAAAAAAGGUAACAAUCUCCCCUCUUUGGGACUCGAUGCAAACGAAUUAU -.....((((((.((((((((((((((.....)))))).(((((.(((.((.......)).))))---)))).....))))......((((......))))...))))))))))...... ( -32.10, z-score = -2.06, R) >droPer1.super_9 1898666 103 - 3637205 CAGUGCCCUCUGCC-UCUGUCGUUUGCCACUUGUAAUUAUU---------UACGACUAAGUGGCC-------CAAAAGGUAAUAAUCUCCCCCUUAAACAGAGAGAAUUAAAACAAUUCU (((......))).(-(((((.....(((((((((((....)---------)))....))))))).-------...((((............))))..))))))((((((.....)))))) ( -22.00, z-score = -2.48, R) >dp4.chrXR_group6 3602811 102 - 13314419 -AGUGCCCUCUGCC-UCUGUCGUUUGCCACUUGUAAUUAUU---------UACGACUAAGUGGCC-------CAAAAGGUAAUAAUCUCCCCCUUAAACAGAGAGAAUUAAAACAAUUCU -............(-(((((.....(((((((((((....)---------)))....))))))).-------...((((............))))..))))))((((((.....)))))) ( -21.30, z-score = -2.37, R) >droAna3.scaffold_13337 11976454 113 + 23293914 -------CUGGGAUGCAUGCCGCUUGCCACUUGUAAUUAUUUUCAGCCUUUACGAGUAAGUGGCCAGAAAGAAAAAAGGUAACAAUCUCCCCUCUUUGGGACCCGAUGCGAGCGAAUUAU -------.............(((((((.((((((((.............))))))))..(.((((..(((((....((((....))))....))))).)).)))...)))))))...... ( -26.32, z-score = 0.16, R) >droEre2.scaffold_4784 18093959 94 - 25762168 -UUGGCCGUUUGUCGUCUGCCGGUUGCCGCUUGUAAUUAUUUUGUGCCUCUGC----AAGUGGCC-------GAAAAGGCAACAAUCUCCCCUCUUUGGGAAUUAU-------------- -..(((.....)))((.((((..((((((((((((...............)))----)))))).)-------))...))))))....((((......)))).....-------------- ( -26.66, z-score = -1.09, R) >droYak2.chr3L 18449172 112 - 24197627 -UUGGCCGUUUGUCGUCUGCCGGUUGCCACUUGUAAUUAUUUUGUGCCUCUGCAAGUAAGUGGCC-------GAAAAGGUAACGAUCUCCCCUCUUUGGGACACGAUGCAAACGAAUUAU -.....((((((.((((((((..((((((((((.......(((((......)))))))))))).)-------))...))))......((((......))))...))))))))))...... ( -32.21, z-score = -1.41, R) >droSec1.super_0 7947898 114 - 21120651 -UUGGCCGUUUGUCGUCUGCCGGUUGCCACUUGUAAUUUUUUUGUGCCUCUGCAAGUAAGUGGCC-----AAAAGAAGGUAACAAUCUCCCCUCUUUGGGACUCGAUGCAAACGAAUUAU -.....((((((.((((.....((((((........((((((((.(((.((.......)).))))-----)))))))))))))....((((......))))...))))))))))...... ( -34.20, z-score = -2.22, R) >droSim1.chr3L 15185028 115 - 22553184 -UUGGCCGUUUGUCGUCUGCCGGUUGCCACUUGUAAUUAUUUUGUGCCUCUGCAAGUAAGUGGCC----AAAAAGAAGGUAACAAUCUCCCCUCUUCGGGACUCGAUGCAAACGAAUUAU -.....((((((.((((((((((((((.....)))))).(((((.(((.((.......)).))))----))))....))))......((((......))))...))))))))))...... ( -32.20, z-score = -1.69, R) >consensus _UUGGCCGUUUGUCGUCUGCCGGUUGCCACUUGUAAUUAUUUUGUGCCUCUGCAAGUAAGUGGCC_______AAAAAGGUAACAAUCUCCCCUCUUUGGGACUCGAUGCAAACGAAUUAU .................((((..((((((((((.......................)))))))).........))..))))......((((......))))................... (-10.94 = -11.04 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:06 2011