
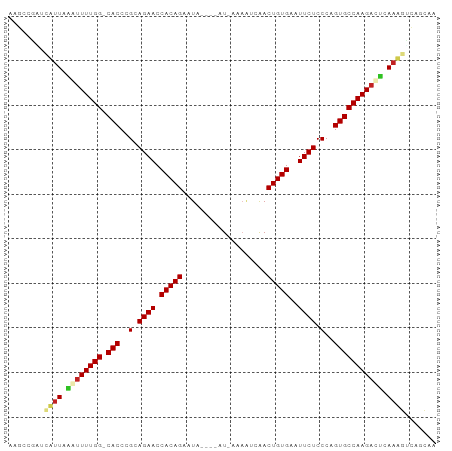
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,836,505 – 15,836,595 |
| Length | 90 |
| Max. P | 0.999850 |

| Location | 15,836,505 – 15,836,595 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 80.52 |
| Shannon entropy | 0.41715 |
| G+C content | 0.44133 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -17.87 |
| Energy contribution | -17.85 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.82 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.10 |
| SVM RNA-class probability | 0.999627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15836505 90 + 24543557 AAGCCGGUUUUUAAGUUUUGG-CACCCGCAGAACCACAGAAUA----AU-AAAGUCAACUGUGAAUUCUCCCAGUGCCAAGACUCAAAGUCAGCAA ..((.(..(((..((((((((-(((..(.((((.(((((....----..-........)))))..)))).)..))))))))))).)))..).)).. ( -27.54, z-score = -4.52, R) >droSim1.chr3L 15181330 90 + 22553184 AAGCCGGUUUUUAAGUUUUGG-CACCCGCAGAACCACAGAAUA----AU-CAAGUCAACUGUGAAUUCUCCCAGUGCCAAGACUCAAAGUCAGCAA ..((.(..(((..((((((((-(((..(.((((.(((((....----..-........)))))..)))).)..))))))))))).)))..).)).. ( -27.54, z-score = -4.41, R) >droSec1.super_0 7944400 90 + 21120651 AAGCCGGUUUUUAAGUUUUGG-CACCCGCAGAACCACAGAAUA----AU-CAAGUCAACUGUGAAUUCUCCCAGUGCCAAGACUCAAAGUCAGCAA ..((.(..(((..((((((((-(((..(.((((.(((((....----..-........)))))..)))).)..))))))))))).)))..).)).. ( -27.54, z-score = -4.41, R) >droYak2.chr3L 18445398 90 + 24197627 GAGCCGUUGAUUAAAUUUUGG-CACCCGCAGAACCACAGAAUA----AU-AAAGUCAACUGUGAAUUCUCCCAGUGCCAAGACUCAAAGUCAGCAG .....(((((((...((((((-(((..(.((((.(((((....----..-........)))))..)))).)..))))))))).....))))))).. ( -25.34, z-score = -4.25, R) >droEre2.scaffold_4784 18090164 85 + 25762168 GAGCUGUUGAUUAAAUUUUGG-CACCCGCAGAACCACAGAAUA----AU-AAAAUCAACUGUGAAUUCUCCCAGUGCCAAGACUCAAAGUC----- ..(((.((((.....((((((-(((..(.((((.(((((....----..-........)))))..)))).)..))))))))).))))))).----- ( -20.04, z-score = -2.75, R) >droAna3.scaffold_13337 11973083 90 - 23293914 AGGCCAUUAUUUCAAUUUUGG-CACCCGCAGAACCACAGAAUA----AU-AAAAUCAACUGUGAAUUCUCCCAGUGCCAAGAUUCAAAGACAGCAG ..((.....(((.((((((((-(((..(.((((.(((((....----..-........)))))..)))).)..))))))))))).)))....)).. ( -23.14, z-score = -4.21, R) >droPer1.super_9 1894045 93 + 3637205 GAGCCGAACAUUCGAUUUUGGGCACCCGCAGAACCACAGAAUAU---AUGAAAACCAACUGUGAAUUCUCCCAGUGCCAAGAGCCAAUGCCAGCAA ..((.(..((((.(.((((.(((((..(.((((.(((((.....---...........)))))..)))).)..))))).))))).)))).).)).. ( -19.09, z-score = -1.21, R) >dp4.chrXR_group6 3598047 92 + 13314419 GAGCCGAACAUUCGAUUUUGG-CACCCGCAGAACCACAGAAUAU---AUGGAAACCAACUGUGAAUUCUCCCAGUGCCAAGAGCCAAUGCCAGCAA ..((.(..((((.(.((((((-(((..(.((((.(((((.....---..(....)...)))))..)))).)..))))))))).).)))).).)).. ( -24.00, z-score = -3.01, R) >droWil1.scaffold_180727 1288972 95 - 2741493 AGCUCAAACAUUAGAUUUUGG-CACCCGCAGAACCACAGAAUAUCGAAUAAGAAUCAACUGUGAAUUCUCCCAGUGCCAAGAUGCAUUGUGAGCAG .((((..(((....(((((((-(((..(.((((.(((((....((......)).....)))))..)))).)..))))))))))....))))))).. ( -28.70, z-score = -4.33, R) >droVir3.scaffold_13049 24412522 86 + 25233164 ------CACAUUACAUUUUGG-CACCCGCAGAACCACAGAAUAUUAUGUAA---UCGACUGUGAAUUCUCCCAGUGCCAAGAUGAAAUGGCAGUAG ------..((((.((((((((-(((..(.((((.(((((...(((....))---)...)))))..)))).)..))))))))))).))))....... ( -26.40, z-score = -4.28, R) >droGri2.scaffold_15110 24319805 86 + 24565398 ------CACAUUACAGUUUGG-CACCCGCAGAACCACAGAAUAUUAUAUAA---CCAACUGUGAAUUCUCCCAGUGCCAAGAUGUAAUGGCAGUAG ------..(((((((.(((((-(((..(.((((.(((((............---....)))))..)))).)..)))))))).)))))))....... ( -26.79, z-score = -4.67, R) >consensus AAGCCGAUCAUUAAAUUUUGG_CACCCGCAGAACCACAGAAUA____AU_AAAAUCAACUGUGAAUUCUCCCAGUGCCAAGACUCAAAGUCAGCAA ........((((.((((((((.(((..(.((((.(((((...................)))))..)))).)..))))))))))).))))....... (-17.87 = -17.85 + -0.02)


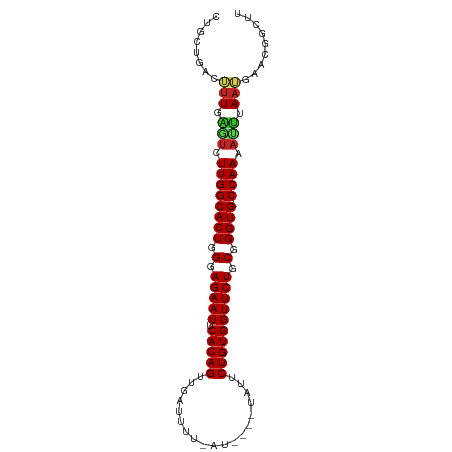
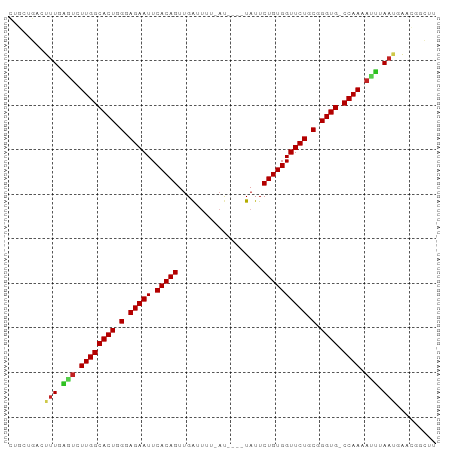
| Location | 15,836,505 – 15,836,595 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 80.52 |
| Shannon entropy | 0.41715 |
| G+C content | 0.44133 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -23.58 |
| Energy contribution | -23.11 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.20 |
| Mean z-score | -4.88 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.57 |
| SVM RNA-class probability | 0.999850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15836505 90 - 24543557 UUGCUGACUUUGAGUCUUGGCACUGGGAGAAUUCACAGUUGACUUU-AU----UAUUCUGUGGUUCUGCGGGUG-CCAAAACUUAAAAACCGGCUU ..((((..(((((((.((((((((.(.(((((.(((((.(((....-.)----))..)))))))))).).))))-)))).)))))))...)))).. ( -37.70, z-score = -5.96, R) >droSim1.chr3L 15181330 90 - 22553184 UUGCUGACUUUGAGUCUUGGCACUGGGAGAAUUCACAGUUGACUUG-AU----UAUUCUGUGGUUCUGCGGGUG-CCAAAACUUAAAAACCGGCUU ..((((..(((((((.((((((((.(.(((((.(((((.(((....-.)----))..)))))))))).).))))-)))).)))))))...)))).. ( -37.70, z-score = -5.75, R) >droSec1.super_0 7944400 90 - 21120651 UUGCUGACUUUGAGUCUUGGCACUGGGAGAAUUCACAGUUGACUUG-AU----UAUUCUGUGGUUCUGCGGGUG-CCAAAACUUAAAAACCGGCUU ..((((..(((((((.((((((((.(.(((((.(((((.(((....-.)----))..)))))))))).).))))-)))).)))))))...)))).. ( -37.70, z-score = -5.75, R) >droYak2.chr3L 18445398 90 - 24197627 CUGCUGACUUUGAGUCUUGGCACUGGGAGAAUUCACAGUUGACUUU-AU----UAUUCUGUGGUUCUGCGGGUG-CCAAAAUUUAAUCAACGGCUC ..((((...((((((.((((((((.(.(((((.(((((.(((....-.)----))..)))))))))).).))))-)))).))))))....)))).. ( -34.80, z-score = -5.13, R) >droEre2.scaffold_4784 18090164 85 - 25762168 -----GACUUUGAGUCUUGGCACUGGGAGAAUUCACAGUUGAUUUU-AU----UAUUCUGUGGUUCUGCGGGUG-CCAAAAUUUAAUCAACAGCUC -----....((((((.((((((((.(.(((((.(((((.((((...-))----))..)))))))))).).))))-)))).)))))).......... ( -30.10, z-score = -4.21, R) >droAna3.scaffold_13337 11973083 90 + 23293914 CUGCUGUCUUUGAAUCUUGGCACUGGGAGAAUUCACAGUUGAUUUU-AU----UAUUCUGUGGUUCUGCGGGUG-CCAAAAUUGAAAUAAUGGCCU ..(((((.(((.(((.((((((((.(.(((((.(((((.((((...-))----))..)))))))))).).))))-)))).))).)))..))))).. ( -32.20, z-score = -4.52, R) >droPer1.super_9 1894045 93 - 3637205 UUGCUGGCAUUGGCUCUUGGCACUGGGAGAAUUCACAGUUGGUUUUCAU---AUAUUCUGUGGUUCUGCGGGUGCCCAAAAUCGAAUGUUCGGCUC ..(((((((((.(.....((((((.(.(((((.(((((...((......---))...)))))))))).).))))))......).)))).))))).. ( -34.30, z-score = -3.35, R) >dp4.chrXR_group6 3598047 92 - 13314419 UUGCUGGCAUUGGCUCUUGGCACUGGGAGAAUUCACAGUUGGUUUCCAU---AUAUUCUGUGGUUCUGCGGGUG-CCAAAAUCGAAUGUUCGGCUC ..(((((((((.(...((((((((.(.(((((.(((((...((......---))...)))))))))).).))))-))))...).)))).))))).. ( -37.70, z-score = -4.49, R) >droWil1.scaffold_180727 1288972 95 + 2741493 CUGCUCACAAUGCAUCUUGGCACUGGGAGAAUUCACAGUUGAUUCUUAUUCGAUAUUCUGUGGUUCUGCGGGUG-CCAAAAUCUAAUGUUUGAGCU ..(((((((.((.((.((((((((.(.(((((.(((((((((.......))))....)))))))))).).))))-)))).)).)).)))..)))). ( -34.40, z-score = -4.18, R) >droVir3.scaffold_13049 24412522 86 - 25233164 CUACUGCCAUUUCAUCUUGGCACUGGGAGAAUUCACAGUCGA---UUACAUAAUAUUCUGUGGUUCUGCGGGUG-CCAAAAUGUAAUGUG------ .......((((.(((.((((((((.(.(((((.(((((...(---((....)))...)))))))))).).))))-)))).))).))))..------ ( -32.50, z-score = -4.83, R) >droGri2.scaffold_15110 24319805 86 - 24565398 CUACUGCCAUUACAUCUUGGCACUGGGAGAAUUCACAGUUGG---UUAUAUAAUAUUCUGUGGUUCUGCGGGUG-CCAAACUGUAAUGUG------ .......(((((((..((((((((.(.(((((.(((((...(---((....)))...)))))))))).).))))-))))..)))))))..------ ( -35.70, z-score = -5.54, R) >consensus CUGCUGACUUUGAGUCUUGGCACUGGGAGAAUUCACAGUUGAUUUU_AU____UAUUCUGUGGUUCUGCGGGUG_CCAAAAUUUAAUGAACGGCUU ........(((.(((.((((((((.(.(((((.(((((...................)))))))))).).)))).)))).))).)))......... (-23.58 = -23.11 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:04 2011