
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,790,427 – 15,790,530 |
| Length | 103 |
| Max. P | 0.791921 |

| Location | 15,790,427 – 15,790,530 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 87.73 |
| Shannon entropy | 0.25875 |
| G+C content | 0.40609 |
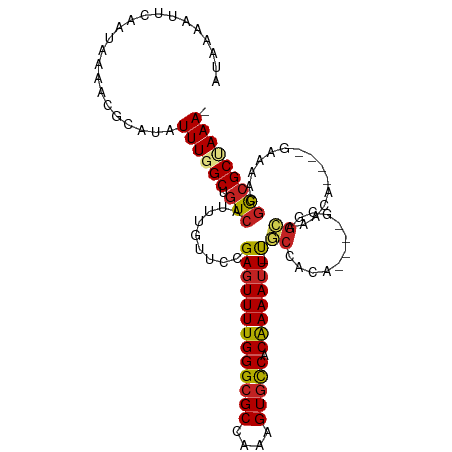
| Mean single sequence MFE | -30.89 |
| Consensus MFE | -16.90 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
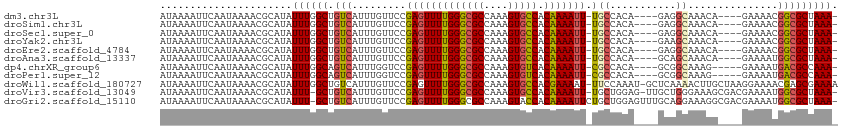
>dm3.chr3L 15790427 103 - 24543557 AUAAAAUUCAAUAAAACGCAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUU-UGCCACA----GAGGCAAACA----GAAAACGGCGCUAAA- ................(((...((((..((((.(((((..((((((((((((((....))))).)))))))-))..)))----))))))..))----)).....))).....- ( -31.50, z-score = -2.94, R) >droSim1.chr3L 15131761 103 - 22553184 AUAAAAUUCAAUAAAACGCAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUU-UGCCACA----GAGGCAAACA----GAAAACGGCGCUAAA- ................(((...((((..((((.(((((..((((((((((((((....))))).)))))))-))..)))----))))))..))----)).....))).....- ( -31.50, z-score = -2.94, R) >droSec1.super_0 7900747 103 - 21120651 AUAAAAUUCAAUAAAACGCAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUU-UGCCACA----GAGGCAAACA----GAAAACGGCGCUAAA- ................(((...((((..((((.(((((..((((((((((((((....))))).)))))))-))..)))----))))))..))----)).....))).....- ( -31.50, z-score = -2.94, R) >droYak2.chr3L 18344309 103 - 24197627 AUAAAAUUCAAUAAAACGCAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUU-UGCCACA----GAAGCAAACA----GAAAACGGCGCUAAA- ................(((...((((..(((..(((((..((((((((((((((....))))).)))))))-))..)))----)).)))..))----)).....))).....- ( -28.10, z-score = -2.17, R) >droEre2.scaffold_4784 18045578 103 - 25762168 AUAAAAUUCAAUAAAACGCAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUU-UGCCACA----GAGGCAAACA----GAAAACGGCGCUAAA- ................(((...((((..((((.(((((..((((((((((((((....))))).)))))))-))..)))----))))))..))----)).....))).....- ( -31.50, z-score = -2.94, R) >droAna3.scaffold_13337 11933091 103 + 23293914 AUAAAAUUCAAUAAAACGCAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUU-UGCCACA----GCAGCAAACA----GAAAAUGGCGCUAAA- ................(((.(((((.(((((....(((..((((((((((((((....))))).)))))))-))..)))----))))).....----..)))))))).....- ( -28.00, z-score = -1.73, R) >dp4.chrXR_group6 13305557 102 + 13314419 AUAAAAUUCAAUAAAACGCAUAUUUGGCAGUCAUUUGGUCCGAGUUUUGGGCGCCAAAGUGUCACAAAAUU-CGCCACA----GCGGCAAAG-----GAAAAUGACGCCAAA- ......................((((((.(((((((..((((((((((((((((....))))).)))))))-)(((...----..)))...)-----)))))))))))))))- ( -36.40, z-score = -4.88, R) >droPer1.super_12 1728906 102 + 2414086 AUAAAAUUCAAUAAAACGCAUAUUUGGCAGUCAUUUGGUCCGAGUUUUGGGCGCCAAAGUGUCACAAAAUU-CGCCACA----GCGGCAAAG-----GAAAAUGACGCCAAA- ......................((((((.(((((((..((((((((((((((((....))))).)))))))-)(((...----..)))...)-----)))))))))))))))- ( -36.40, z-score = -4.88, R) >droWil1.scaffold_180727 1517253 111 - 2741493 AUAAAAUUCAAUAAAACGCAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACGAAAAU-UUCCAAAU-GCUCAAAACUUGCUAAGGAAAACGAGCGAAAA .................((.......)).....(((((((....((((((((((....))))).))))).(-((((....-((.........))...)))))..))))))).. ( -22.20, z-score = -0.06, R) >droVir3.scaffold_13049 2983288 109 + 25233164 AUAAAAUUCAAUAAAACGCAUAUUU-GCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUU-UGCUGGAG-UUGCUGGGAAAGCGACGAAAAUGGCGCUAAA- .................(((....)-)).(((((((..((((((((((((((((....))))).)))))))-)...)))(-(((((.....))))))..)))))))......- ( -33.30, z-score = -2.02, R) >droGri2.scaffold_15110 21292490 111 - 24565398 AUAAAAUUCAAUAAAACGCAUAUUU-GCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUACCACAAAAUUCUGCUGGAGUUUGCAGGAAAGGCGACGAAAAUGGCGCUAAA- .(((((((((((((((((((....)-)).))..))))))..))))))))(((((((.............((((((........))))))..(....).....)))))))...- ( -29.40, z-score = -1.17, R) >consensus AUAAAAUUCAAUAAAACGCAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUU_UGCCACA____GCGGCAAACA____GAAAACGGCGCUAAA_ ......................((((((.(((..........((((((((((((....))))).))))))).(((...........)))..............))))))))). (-16.90 = -17.32 + 0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:30:01 2011