| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,783,840 – 15,783,938 |
| Length | 98 |
| Max. P | 0.965321 |

| Location | 15,783,840 – 15,783,938 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 66.28 |
| Shannon entropy | 0.53517 |
| G+C content | 0.36032 |
| Mean single sequence MFE | -17.78 |
| Consensus MFE | -10.36 |
| Energy contribution | -10.60 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.595684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
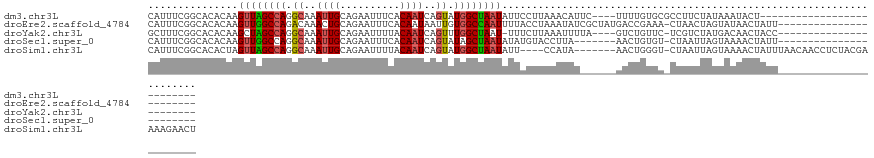
>dm3.chr3L 15783840 98 + 24543557 CAUUUCGGCACACAAGUUAGCCAGGCAAAUUGCAGAAUUUCACAAUCAGUAUGGCUAAUAUUCCUUAAACAUUC----UUUUGUGCGCCUUCUAUAAAUACU-------------------------- ......(((.(((((((((((((.((..((((..........))))..)).))))))))...............----..))))).))).............-------------------------- ( -21.82, z-score = -2.86, R) >droEre2.scaffold_4784 18039017 104 + 25762168 CAUUUCGGCACACAAGUUGGCCAGACAAACUGCAGAAUUUCACAAUAAUUGUGGCCAAUUUUACCUAAAUAUCGCUAUGACCGAAA-CUAACUAGUAUAACUAUU----------------------- ..((((((.....(((((((((((.....)))........((((.....))))))))))))..........((.....))))))))-..................----------------------- ( -16.30, z-score = -0.63, R) >droYak2.chr3L 18336917 99 + 24197627 GCUUUCGGCACACAAGCUAGCCAGGCAAAUUGCAGAAUUUUACAAUCAGUUUGGCUAAU-UUUCUUAAAUUUUA----GUCUGUUC-UCGUCUAUGACAACUACC----------------------- ((.....))........(((((((((..((((..........))))..)))))))))..-............((----((.(((.(-........))))))))..----------------------- ( -16.80, z-score = -0.27, R) >droSec1.super_0 7894206 97 + 21120651 CAUUUCGGCACACAAGUUGGCCAGGCAAAUUGCAGAAUUUCACAAUCAGUAUAGCUAAUAUAUGUACCUUA-------AACUGUGU-CUAAUUAGUAAAACUAUU----------------------- ......((((((...((((((...((..((((..........))))..))...))))))....((......-------.)))))))-).................----------------------- ( -12.70, z-score = 1.02, R) >droSim1.chr3L 15124513 116 + 22553184 CAUUUCGGCACACUAGUUAGCCAGGCAAAUUGCAGAAUUUUACAAUCAGUAUGGCUAAUAUU----CCAUA-------AACUGGGU-CUAAUUAGUAAAACUAUUUAACAACCUCUACGAAAAGAACU ..(((((........((((((((.((..((((..........))))..)).))))))))...----.....-------....((((-.(((.(((.....))).)))...))))...)))))...... ( -21.30, z-score = -1.53, R) >consensus CAUUUCGGCACACAAGUUAGCCAGGCAAAUUGCAGAAUUUCACAAUCAGUAUGGCUAAUAUUUCUUAAAUAU______AACUGUGC_CUAACUAGUAAAACUAUU_______________________ ...............((((((((.((..((((..........))))..)).))))))))..................................................................... (-10.36 = -10.60 + 0.24)
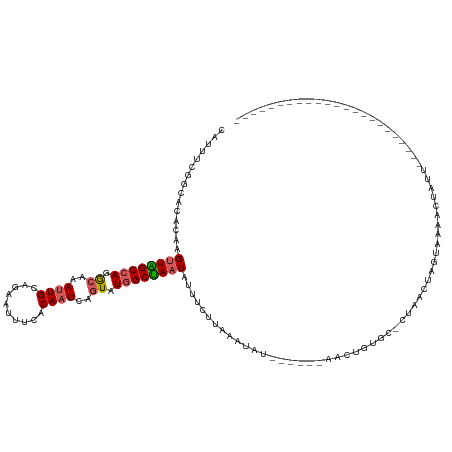
| Location | 15,783,840 – 15,783,938 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 128 |
| Reading direction | reverse |
| Mean pairwise identity | 66.28 |
| Shannon entropy | 0.53517 |
| G+C content | 0.36032 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -13.08 |
| Energy contribution | -12.52 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15783840 98 - 24543557 --------------------------AGUAUUUAUAGAAGGCGCACAAAA----GAAUGUUUAAGGAAUAUUAGCCAUACUGAUUGUGAAAUUCUGCAAUUUGCCUGGCUAACUUGUGUGCCGAAAUG --------------------------.............(((((((((..----.(((((((...)))))))(((((..(.((((((........)))))).)..)))))...)))))))))...... ( -28.20, z-score = -3.25, R) >droEre2.scaffold_4784 18039017 104 - 25762168 -----------------------AAUAGUUAUACUAGUUAG-UUUCGGUCAUAGCGAUAUUUAGGUAAAAUUGGCCACAAUUAUUGUGAAAUUCUGCAGUUUGUCUGGCCAACUUGUGUGCCGAAAUG -----------------------..(((.....)))....(-((((((((((((...(((....)))...(((((((.....(((((........))))).....))))))).))))).)))))))). ( -25.10, z-score = -1.46, R) >droYak2.chr3L 18336917 99 - 24197627 -----------------------GGUAGUUGUCAUAGACGA-GAACAGAC----UAAAAUUUAAGAAA-AUUAGCCAAACUGAUUGUAAAAUUCUGCAAUUUGCCUGGCUAGCUUGUGUGCCGAAAGC -----------------------(((((((.((......))-.)))....----..............-.(((((((..(.(((((((......))))))).)..)))))))......))))...... ( -22.60, z-score = -1.36, R) >droSec1.super_0 7894206 97 - 21120651 -----------------------AAUAGUUUUACUAAUUAG-ACACAGUU-------UAAGGUACAUAUAUUAGCUAUACUGAUUGUGAAAUUCUGCAAUUUGCCUGGCCAACUUGUGUGCCGAAAUG -----------------------....(((((.....((((-((...)))-------)))((((((((..((.((((..(.((((((........)))))).)..)))).))..))))))))))))). ( -22.70, z-score = -1.94, R) >droSim1.chr3L 15124513 116 - 22553184 AGUUCUUUUCGUAGAGGUUGUUAAAUAGUUUUACUAAUUAG-ACCCAGUU-------UAUGG----AAUAUUAGCCAUACUGAUUGUAAAAUUCUGCAAUUUGCCUGGCUAACUAGUGUGCCGAAAUG ..(((.(((((((((((((..(((.(((.....))).))))-)))...))-------)))))----))..(((((((..(.(((((((......))))))).)..)))))))..........)))... ( -24.90, z-score = -1.20, R) >consensus _______________________AAUAGUUUUACUAGUUAG_ACACAGUC______AUAUUUAAGAAAUAUUAGCCAUACUGAUUGUGAAAUUCUGCAAUUUGCCUGGCUAACUUGUGUGCCGAAAUG ......................................................................(((((((....(((((((......)))))))....)))))))................ (-13.08 = -12.52 + -0.56)
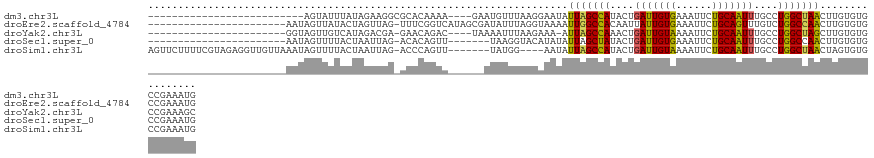
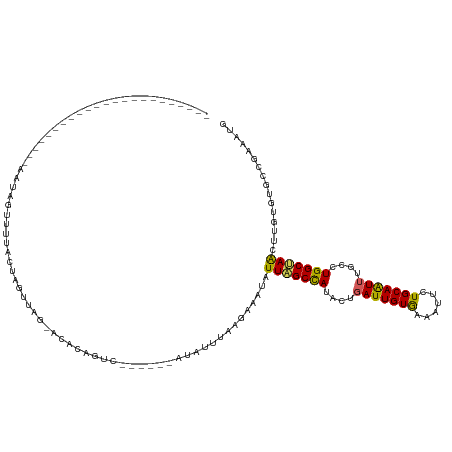
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:29:59 2011