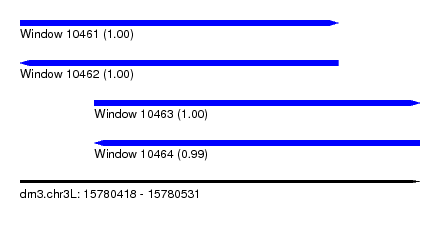
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,780,418 – 15,780,531 |
| Length | 113 |
| Max. P | 0.998025 |

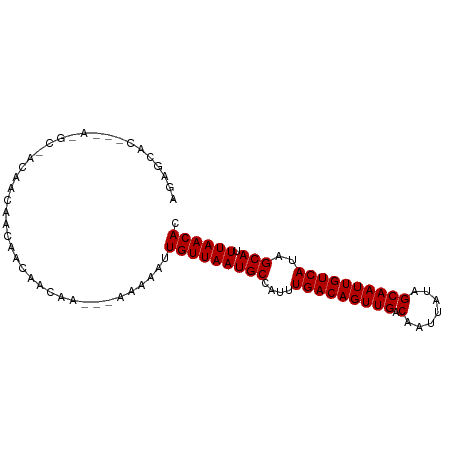
| Location | 15,780,418 – 15,780,508 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 77.75 |
| Shannon entropy | 0.44363 |
| G+C content | 0.32954 |
| Mean single sequence MFE | -18.50 |
| Consensus MFE | -16.17 |
| Energy contribution | -16.17 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15780418 90 + 24543557 AGAGCACUGUA-GCAACAGCAACAACAACAA---AGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACAC .......(((.-((....)).))).......---......(((((((((....(((((((((.(.......))))))))))..))).)))))). ( -19.30, z-score = -2.17, R) >droSim1.chr3L 15121144 93 + 22553184 AGAGCACUGUA-GCAACAGCAACAACAACAACAAAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACAC .......(((.-((....)).)))................(((((((((....(((((((((.(.......))))))))))..))).)))))). ( -19.30, z-score = -2.14, R) >droSec1.super_0 7890861 87 + 21120651 AGAGCACUGUA-GC---AACAGCAACAACAA---AGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACAC .......(((.-((---....)).)))....---......(((((((((....(((((((((.(.......))))))))))..))).)))))). ( -19.30, z-score = -2.16, R) >droEre2.scaffold_4784 18035538 87 + 25762168 AGAGCACUGUA-GC---AGCAACAACAACAA---AGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACAC ...(((((((.-..---.(((..(((((...---.....)))))..)))......)))))((((((((.....))))))))..))......... ( -18.00, z-score = -1.64, R) >droAna3.scaffold_13337 11922972 81 - 23293914 ----------AGGGCACACUGGCUACAUCAA---AAAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACAC ----------..(((......))).......---......(((((((((....(((((((((.(.......))))))))))..))).)))))). ( -18.50, z-score = -2.22, R) >droWil1.scaffold_180727 1525566 73 + 2741493 ----------AGUGCAAAAGGGC-----------AAAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACAC ----------..(((......))-----------).....(((((((((....(((((((((.(.......))))))))))..))).)))))). ( -18.40, z-score = -2.56, R) >droVir3.scaffold_13049 22259626 74 + 25233164 ----------AGAGCAACAACAG-------A---AAAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAAAGCAUUUAACAC ----------.............-------.---......(((((((((..(((((((((((.(.......))))))))))))))).)))))). ( -17.90, z-score = -3.83, R) >droMoj3.scaffold_6680 16866152 93 + 24764193 AGAGCACCAAAUACAACAACAACAAAAACAACA-AAAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAAAGCAUUUAACAC .................................-......(((((((((..(((((((((((.(.......))))))))))))))).)))))). ( -17.90, z-score = -3.65, R) >droGri2.scaffold_8832 2487 73 - 5417 --------------------AAAGCAACAGACA-AAAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAAAGCAUUUAACAC --------------------.............-......(((((((((..(((((((((((.(.......))))))))))))))).)))))). ( -17.90, z-score = -3.76, R) >consensus AGAGCAC___A_GC_ACAACAACAACAACAA___AAAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACAC ........................................(((((((((....(((((((((.(.......))))))))))..))).)))))). (-16.17 = -16.17 + 0.00)

| Location | 15,780,418 – 15,780,508 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 77.75 |
| Shannon entropy | 0.44363 |
| G+C content | 0.32954 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -14.73 |
| Energy contribution | -14.73 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.997965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15780418 90 - 24543557 GUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCU---UUGUUGUUGUUGCUGUUGC-UACAGUGCUCU .........((..(((((((((...)))))))))(((((..(((((((.(((((((....---..))))))).)))))))..-.)))))))... ( -26.60, z-score = -3.79, R) >droSim1.chr3L 15121144 93 - 22553184 GUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUUUGUUGUUGUUGUUGCUGUUGC-UACAGUGCUCU .........((..(((((((((...)))))))))(((((..(((((((.(((((((.........))))))).)))))))..-.)))))))... ( -25.60, z-score = -3.01, R) >droSec1.super_0 7890861 87 - 21120651 GUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCU---UUGUUGUUGCUGUU---GC-UACAGUGCUCU .........((..(((((((((...)))))))))(((((..(((((((.((((((.....---)))))).)))))))---..-.)))))))... ( -25.30, z-score = -3.59, R) >droEre2.scaffold_4784 18035538 87 - 25762168 GUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCU---UUGUUGUUGUUGCU---GC-UACAGUGCUCU .........((..(((((((((...)))))))))(((((....(((((.(((((((....---..))))))).))))---).-.)))))))... ( -22.50, z-score = -2.49, R) >droAna3.scaffold_13337 11922972 81 + 23293914 GUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUUU---UUGAUGUAGCCAGUGUGCCCU---------- .((((((.((((((((((.((((.......).))).)))))..)))))))))))......---.....................---------- ( -15.80, z-score = -0.47, R) >droWil1.scaffold_180727 1525566 73 - 2741493 GUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUUU-----------GCCCUUUUGCACU---------- .((((((.((((((((((.((((.......).))).)))))..))))))))))).....(-----------((......)))..---------- ( -16.90, z-score = -2.45, R) >droVir3.scaffold_13049 22259626 74 - 25233164 GUGUUAAAUGCUUUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUUU---U-------CUGUUGUUGCUCU---------- ......((((((((((((.((((.......).))).))))))..))))))((((((....---.-------..)))))).....---------- ( -15.80, z-score = -2.30, R) >droMoj3.scaffold_6680 16866152 93 - 24764193 GUGUUAAAUGCUUUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUUU-UGUUGUUUUUGUUGUUGUUGUAUUUGGUGCUCU (..((((((((.((((((((((...))))))))))......((..(((..((((((....-.))))))..)))..))...))))))))..)... ( -27.20, z-score = -4.77, R) >droGri2.scaffold_8832 2487 73 + 5417 GUGUUAAAUGCUUUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUUU-UGUCUGUUGCUUU-------------------- .((((((.((((((((((.((((.......).))).))))))..))))))))))......-.............-------------------- ( -15.70, z-score = -2.02, R) >consensus GUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUUU___UUGUUGUUGUUGUUGU_GC_U___GUGCUCU .((((((.((((.(((((.((((.......).))).)))))...))))))))))........................................ (-14.73 = -14.73 + -0.00)

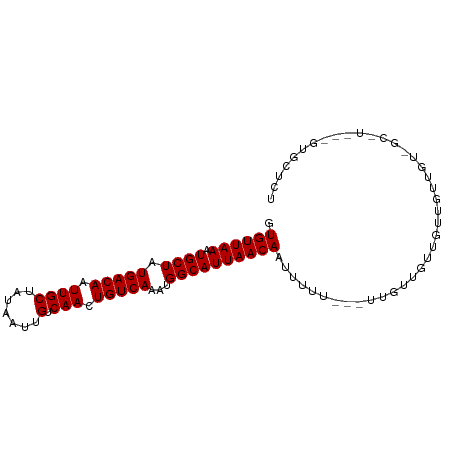
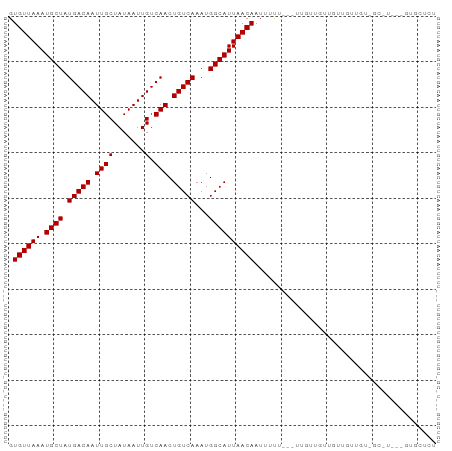
| Location | 15,780,439 – 15,780,531 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.31 |
| Shannon entropy | 0.47206 |
| G+C content | 0.34445 |
| Mean single sequence MFE | -19.57 |
| Consensus MFE | -15.19 |
| Energy contribution | -15.19 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.19 |
| SVM RNA-class probability | 0.997830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15780439 92 + 24543557 --------CAACAACA-AAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGCUGAGCACUAUACGAAAU------------- --------........-.((...((((.(((((....(((((((((.(.......))))))))))..)))))....))))...))................------------- ( -16.50, z-score = -1.63, R) >droSim1.chr3L 15121168 92 + 22553184 --------CAACAACA-AAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGCUGAGCACUAUACGAAAU------------- --------........-.((...((((.(((((....(((((((((.(.......))))))))))..)))))....))))...))................------------- ( -16.50, z-score = -1.63, R) >droSec1.super_0 7890879 82 + 21120651 --------CAACAACA-AAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGA-GAGAAAU---------------------- --------........-.......(((((((((....(((((((((.(.......))))))))))..))).))))))(....).-.......---------------------- ( -16.60, z-score = -3.57, R) >droYak2.chr3L 18333453 95 + 24197627 -----CAGCAACAACA-AAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGCUGAGCACUAUACGAAAU------------- -----((((.......-((....))((((((((....(((((((((.(.......))))))))))..))).)))))......))))...............------------- ( -20.70, z-score = -2.75, R) >droEre2.scaffold_4784 18035556 92 + 25762168 --------CAACAACA-AAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGCUGAGCACUAUACGAAAU------------- --------........-.((...((((.(((((....(((((((((.(.......))))))))))..)))))....))))...))................------------- ( -16.50, z-score = -1.63, R) >droAna3.scaffold_13337 11922984 96 - 23293914 --------CUACAUCA-AAAAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGUUGAGCACUAGGCGAAAAAUAA--------- --------.....(((-(......(((((((((....(((((((((.(.......))))))))))..))).))))))......))))((.....)).........--------- ( -20.30, z-score = -2.26, R) >dp4.chrXR_group6 13297703 109 - 13314419 -----GAGCAACAGCAGCAAAAAAGGUUAAUGCCAUUUGACAGUUGACAAUUAUAACAAUUGUCAUAGCAUUUAACAGGAGAACUGAACUGUUAGGGGGAGGACUAGGCAAAAG -----..((....)).((......((((....((.(((((((((((((((((.....)))))))...........(((.....))))))))))))).))..))))..))..... ( -26.40, z-score = -3.02, R) >droPer1.super_12 1720812 109 - 2414086 -----GAGCAACAGCAGCAAAAAAGGUUAAUGCCAUUUGACAGUUGACAAUUAUAACAAUUGUCAUAGCAUUUAACAGGAGAACUGAACUGUUAGGGGGAGGACUAGGCAAAAG -----..((....)).((......((((....((.(((((((((((((((((.....)))))))...........(((.....))))))))))))).))..))))..))..... ( -26.40, z-score = -3.02, R) >droWil1.scaffold_180727 1525570 83 + 2741493 --------CAAAAGGG-CAAAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGACCAGUAUGAGAA---------------------- --------.....((.-.......(((((((((....(((((((((.(.......))))))))))..))).))))))...))..........---------------------- ( -16.70, z-score = -1.73, R) >droVir3.scaffold_13049 22259630 86 + 25233164 ------CAACAACAG--AAAAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAAAGCAUUUAACACGAGCAGAGUGCAAAAC-------------------- ------.........--.......(((((((((..(((((((((((.(.......))))))))))))))).))))))...((.....)).....-------------------- ( -19.40, z-score = -2.90, R) >droMoj3.scaffold_6680 16866168 91 + 24764193 CAACAACAAAAACAAC-AAAAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAAAGCAUUUAACACGAGCAGAGUGCAAA---------------------- ................-.......(((((((((..(((((((((((.(.......))))))))))))))).))))))...((.....))...---------------------- ( -19.40, z-score = -2.72, R) >droGri2.scaffold_8832 2491 85 - 5417 --------CAACAGAC-AAAAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAAAGCAUUUAACACGAGCAGAGUGCAAGAG-------------------- --------........-.......(((((((((..(((((((((((.(.......))))))))))))))).))))))...((.....)).....-------------------- ( -19.40, z-score = -2.98, R) >consensus ________CAACAACA_AAAAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAACUGAGCAAUAU__G__A______________ .........................((((((((....(((((((((..........)))))))))..))).)))))...................................... (-15.19 = -15.19 + 0.00)

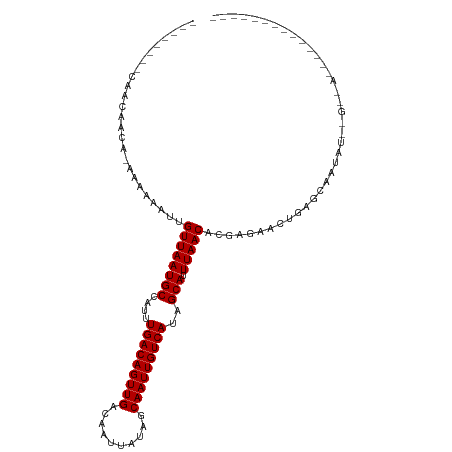
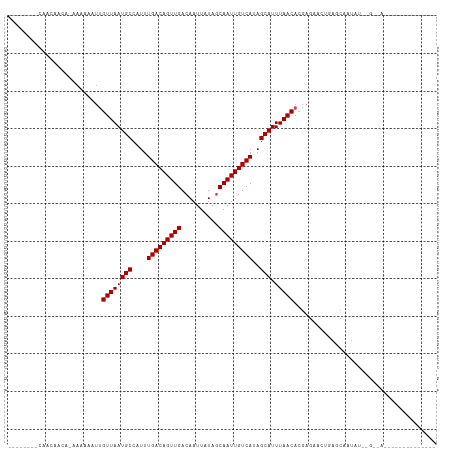
| Location | 15,780,439 – 15,780,531 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.31 |
| Shannon entropy | 0.47206 |
| G+C content | 0.34445 |
| Mean single sequence MFE | -18.95 |
| Consensus MFE | -13.78 |
| Energy contribution | -13.81 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.994857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15780439 92 - 24543557 -------------AUUUCGUAUAGUGCUCAGCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUU-UGUUGUUG-------- -------------...............((((.....((((((.((((((((((.((((.......).))).)))))..))))))))))).......-.))))...-------- ( -18.02, z-score = -1.40, R) >droSim1.chr3L 15121168 92 - 22553184 -------------AUUUCGUAUAGUGCUCAGCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUU-UGUUGUUG-------- -------------...............((((.....((((((.((((((((((.((((.......).))).)))))..))))))))))).......-.))))...-------- ( -18.02, z-score = -1.40, R) >droSec1.super_0 7890879 82 - 21120651 ----------------------AUUUCUC-UCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUU-UGUUGUUG-------- ----------------------.......-............((((((((((((.((((.......).))).)))))..)))))))((((((.....-.)))))).-------- ( -16.40, z-score = -3.69, R) >droYak2.chr3L 18333453 95 - 24197627 -------------AUUUCGUAUAGUGCUCAGCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUU-UGUUGUUGCUG----- -------------............((.((((.....((((((.((((((((((.((((.......).))).)))))..))))))))))).......-.))))..))..----- ( -20.72, z-score = -1.85, R) >droEre2.scaffold_4784 18035556 92 - 25762168 -------------AUUUCGUAUAGUGCUCAGCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUU-UGUUGUUG-------- -------------...............((((.....((((((.((((((((((.((((.......).))).)))))..))))))))))).......-.))))...-------- ( -18.02, z-score = -1.40, R) >droAna3.scaffold_13337 11922984 96 + 23293914 ---------UUAUUUUUCGCCUAGUGCUCAACUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUUUU-UGAUGUAG-------- ---------...............(((((((......((((((.((((((((((.((((.......).))).)))))..)))))))))))......)-))).))).-------- ( -20.00, z-score = -2.22, R) >dp4.chrXR_group6 13297703 109 + 13314419 CUUUUGCCUAGUCCUCCCCCUAACAGUUCAGUUCUCCUGUUAAAUGCUAUGACAAUUGUUAUAAUUGUCAACUGUCAAAUGGCAUUAACCUUUUUUGCUGCUGUUGCUC----- ....................((((((..((((......(((((.((((((((((.(((.((....)).))).)))))..)))))))))).......))))))))))...----- ( -21.92, z-score = -2.87, R) >droPer1.super_12 1720812 109 + 2414086 CUUUUGCCUAGUCCUCCCCCUAACAGUUCAGUUCUCCUGUUAAAUGCUAUGACAAUUGUUAUAAUUGUCAACUGUCAAAUGGCAUUAACCUUUUUUGCUGCUGUUGCUC----- ....................((((((..((((......(((((.((((((((((.(((.((....)).))).)))))..)))))))))).......))))))))))...----- ( -21.92, z-score = -2.87, R) >droWil1.scaffold_180727 1525570 83 - 2741493 ----------------------UUCUCAUACUGGUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUUUG-CCCUUUUG-------- ----------------------..........(((..((((((.((((((((((.((((.......).))).)))))..)))))))))))......)-))......-------- ( -17.30, z-score = -2.45, R) >droVir3.scaffold_13049 22259630 86 - 25233164 --------------------GUUUUGCACUCUGCUCGUGUUAAAUGCUUUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUUUU--CUGUUGUUG------ --------------------.....((((.......))))..((((((((((((.((((.......).))).))))))..))))))((((((.....--..)))))).------ ( -17.90, z-score = -1.90, R) >droMoj3.scaffold_6680 16866168 91 - 24764193 ----------------------UUUGCACUCUGCUCGUGUUAAAUGCUUUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUUUU-GUUGUUUUUGUUGUUG ----------------------...(((....((....))....))).((((((((((...))))))))))......((..(((..((((((.....-))))))..)))..)). ( -20.60, z-score = -2.23, R) >droGri2.scaffold_8832 2491 85 + 5417 --------------------CUCUUGCACUCUGCUCGUGUUAAAUGCUUUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUUUU-GUCUGUUG-------- --------------------.....(((..(......((((((.((((((((((.((((.......).))).))))))..)))))))))).......-)..)))..-------- ( -16.62, z-score = -2.04, R) >consensus ______________U__C__AUAGUGCUCAGCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUUUU_UGUUGUUG________ .....................................((((((.((((.(((((.((((.......).))).)))))...))))))))))........................ (-13.78 = -13.81 + 0.03)


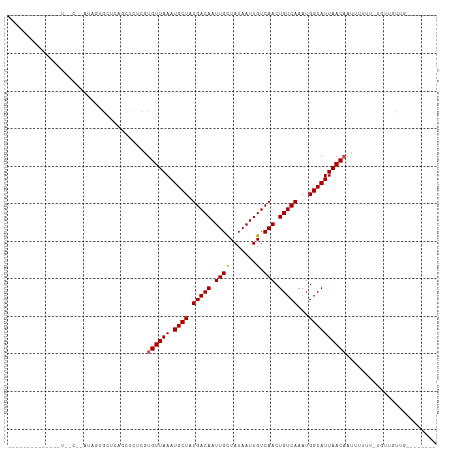
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:29:58 2011