| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,777,937 – 15,778,030 |
| Length | 93 |
| Max. P | 0.845053 |

| Location | 15,777,937 – 15,778,030 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 78.78 |
| Shannon entropy | 0.42172 |
| G+C content | 0.49773 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -18.77 |
| Energy contribution | -20.47 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845053 |
| Prediction | RNA |
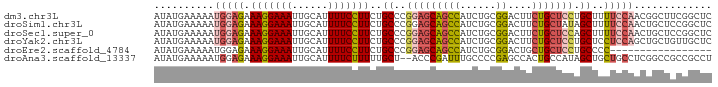
Download alignment: ClustalW | MAF
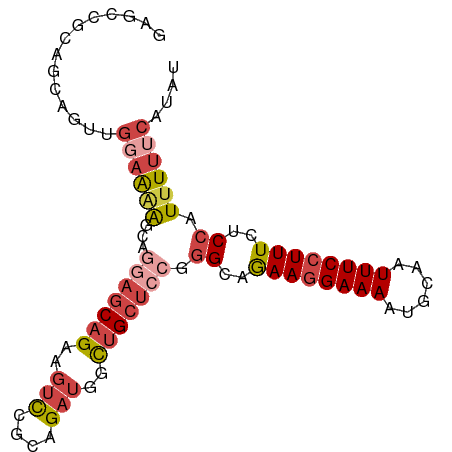
>dm3.chr3L 15777937 93 + 24543557 GAGCCGAAGCCGUUGGAAAAGCAGGAGCAGAAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUUUUCAUAU .....((((....((((...((.(((((((..(((....)))..)))))))..)).((((((((......)))))))).))))..)))).... ( -32.20, z-score = -1.93, R) >droSim1.chr3L 15118647 93 + 22553184 GAGCCGGAGCAGUUGGAAAAGCUAUAGCAGAAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUUUUCAUAU (((((((((((((((((...((....)).....))))......))))))))))...((((((((......)))))))))))............ ( -31.20, z-score = -1.95, R) >droSec1.super_0 7888366 93 + 21120651 GAGCCGGAGCAGUUGGAAAAGCUGGAGCAGAAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUUUUCAUAU .....((((.....(((((..(((((((((..(((....)))..)))))))))(((.........)))..)))))...))))........... ( -32.50, z-score = -1.56, R) >droYak2.chr3L 18330946 93 + 24197627 GAGCAACAGCAGCUGGAGGAGCAGGAGCAGAAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUUUUCAUAU ..((....))...(((((..((.(((((((..(((....)))..)))))))..)).((((((((......))))))))))))).......... ( -34.50, z-score = -2.33, R) >droEre2.scaffold_4784 18033065 76 + 25762168 -----------------GGGGCAGGAGCAGCAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUUUUCAUAU -----------------(((((.((((((((.(((....))).))))))))..)).((((((((......)))))))).)))........... ( -30.50, z-score = -2.88, R) >droAna3.scaffold_13337 11920302 91 - 23293914 AGGCGGCGGCCGAGGCAGCAGCUAUGGCAGUGGCUCGGGGCAAAUCGGGU--AGCAAAAAGAAAAUGCAAUUUCCUUUCUCCAUUUUUCAUAU .(((.((.((((.(((....))).)))).)).))).((((.(((..((((--.(((.........))).))))..)))))))........... ( -23.80, z-score = 0.65, R) >consensus GAGCCGCAGCAGUUGGAAAAGCAGGAGCAGAAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUUUUCAUAU ..............((((((...(((((((..(((....)))..))))))).((..((((((((......))))))))..)).)))))).... (-18.77 = -20.47 + 1.70)


| Location | 15,777,937 – 15,778,030 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 78.78 |
| Shannon entropy | 0.42172 |
| G+C content | 0.49773 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -16.68 |
| Energy contribution | -16.60 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.818908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
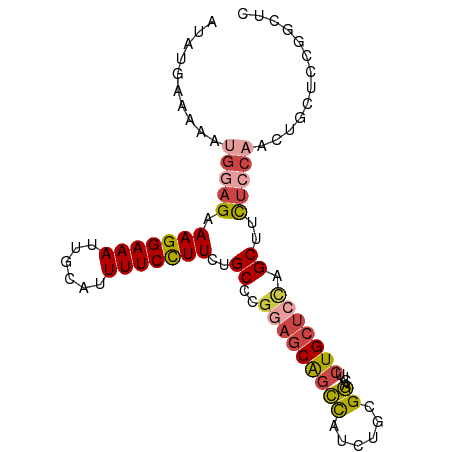
>dm3.chr3L 15777937 93 - 24543557 AUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUUCUGCUCCUGCUUUUCCAACGGCUUCGGCUC ..........(((((((((((((......)))))))..((..(((((((...((....))...))))))).)).))))))..(((....))). ( -30.60, z-score = -2.04, R) >droSim1.chr3L 15118647 93 - 22553184 AUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUUCUGCUAUAGCUUUUCCAACUGCUCCGGCUC ...........(.((((.(((((......))))))))).)(((((((((......((((....))))....(......)..)))))))))... ( -28.60, z-score = -1.92, R) >droSec1.super_0 7888366 93 - 21120651 AUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUUCUGCUCCAGCUUUUCCAACUGCUCCGGCUC ...........(.((((.(((((......))))))))).)(((((((((......((((((....).))).))........)))))))))... ( -29.84, z-score = -1.96, R) >droYak2.chr3L 18330946 93 - 24197627 AUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUUCUGCUCCUGCUCCUCCAGCUGCUGUUGCUC ...........((((((.(((((......))))))))).)).(((((((......((((....))))..)))))))..((((....))))... ( -28.60, z-score = -1.52, R) >droEre2.scaffold_4784 18033065 76 - 25762168 AUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUGCUGCUCCUGCCCC----------------- ...........((((((.(((((......))))))))).)).((((((((..((....))..))))))))......----------------- ( -26.70, z-score = -2.91, R) >droAna3.scaffold_13337 11920302 91 + 23293914 AUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCUUUUUGCU--ACCCGAUUUGCCCCGAGCCACUGCCAUAGCUGCUGCCUCGGCCGCCGCCU ..........(((..((((((((......))))))))..))--)........((.(((((.(((.((....)).).)).)))))..))..... ( -18.50, z-score = 0.65, R) >consensus AUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUUCUGCUCCAGCUUCUCCAACUGCUCCGGCUC ...........((((((.(((((......))))))))).)).(((((((((......))....)))))))....................... (-16.68 = -16.60 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:29:55 2011