
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,748,134 – 15,748,245 |
| Length | 111 |
| Max. P | 0.868020 |

| Location | 15,748,134 – 15,748,227 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 62.79 |
| Shannon entropy | 0.73346 |
| G+C content | 0.47323 |
| Mean single sequence MFE | -25.75 |
| Consensus MFE | -6.43 |
| Energy contribution | -6.44 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
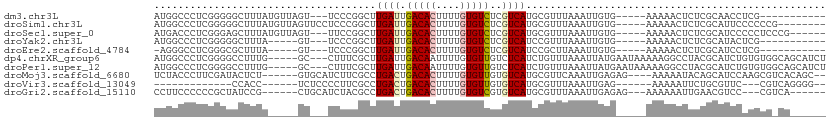
>dm3.chr3L 15748134 93 - 24543557 AUGGCCCUCGGGGGCUUUAUGUUAGU---UCCCGGCUUGAUUGACACUUUUGUGUCUCGUCAUGCGUUUAAAUUGUG-----AAAAACUCUCGCAACCUCG----------- ..((....((((((((.......)))---)))))((.((((.(((((....)))))..)))).)).......(((((-----(.......))))))))...----------- ( -29.20, z-score = -3.17, R) >droSim1.chr3L 15088149 99 - 22553184 AUGGCCCUCGGGGGCUUUAUGUUAGUUCCUCCCGGCUUGAUUGACACUUUUGUGUCUCGUCAUGCGUUUAAAUUGUG-----AAAAACUCUCGCAUUCCCCCCG-------- ........((((((....................((.((((.(((((....)))))..)))).))........((((-----(.......)))))...))))))-------- ( -27.80, z-score = -2.38, R) >droSec1.super_0 7858870 98 - 21120651 AUGACCCUCGGGAGCUUUAUGUUAGU---UUCCGGCUUGAUUGACACUUUUGUGUCUCGUCAUGCGUUUAAAUUGUG-----AAAAACUCUCGCAUCCCCCUCCCG------ ........((((((((.......)))---)))))((.((((.(((((....)))))..)))).))........((((-----(.......)))))...........------ ( -26.00, z-score = -3.10, R) >droYak2.chr3L 18299855 88 - 24197627 AUGGCCCUCGGGGGCUUUA-----GU---UCCCGGCUUGAUUGACACUUUUGUGUCUCGUCAUCCGUUUAAAUUGUG-----AAAAACUCUCGCAUACUCG----------- ((((...((((((((....-----))---))))))..((((.(((((....)))))..)))).))))......((((-----(.......)))))......----------- ( -25.10, z-score = -2.39, R) >droEre2.scaffold_4784 18003624 87 - 25762168 -AGGGCCUCGGGCGCUUUA-----GU---UCCCGGCUUGAUUGACACUUUUGUGUCUCGUCAUCCGCUUAAAUUGUG-----AAAAACUCUCGCAUCCUCG----------- -.((((...(((.((....-----))---.)))((..((((.(((((....)))))..)))).))))))....((((-----(.......)))))......----------- ( -24.30, z-score = -1.88, R) >dp4.chrXR_group6 13271012 104 + 13314419 AUGGCCCUCGGGGCCUUUG-----GC---CUUUCGCUUGAUUGACAAUUUUGUGUUGUCUCAUCUGUUUAAAUUAUGAAUAAAAAGGCCUACGCAUCUGUGUGGCAGCAUCU ..(((((...)))))...(-----((---((((.....(((.((((((.....))))))..)))((((((.....)))))).)))))))(((((....)))))......... ( -30.00, z-score = -1.65, R) >droPer1.super_12 1694063 104 + 2414086 AUGGCCCUCGGGGCCUUUG-----GC---CUUUCGCUUGAUUGACAAUUUUGUGUUGUCUCAUCUGUUUAAAUUAUGAAUAAAAAGGCCUACGCAUCUGUGUGGCAGCAUCU ..(((((...)))))...(-----((---((((.....(((.((((((.....))))))..)))((((((.....)))))).)))))))(((((....)))))......... ( -30.00, z-score = -1.65, R) >droMoj3.scaffold_6680 7859977 100 + 24764193 UCUACCCUUCGAUACUCU------GUGCAUCUUCGCCUGACUGACACUUUUGUGUUGUGUCAUGCGUUCAAAUUGAGAG----AAAAAUACAGCAUCCAAGCGUCACAGC-- ................((------((((.(((((((.((((((((((....)))))).)))).))).((.....)))))----)........((......))).))))).-- ( -22.50, z-score = -0.59, R) >droVir3.scaffold_13049 2939705 82 + 25233164 -------------CCACC------UCUCCCCUUCGCCUGACUGACACUUUUGUGUUGUGUCAUGCGUUUAAAUUGAG------AAAAAUUCUGCGUUC---CGUCAGGGG-- -------------.....------...(((((.(((.((((((((((....)))))).)))).))).........((------(.....)))......---....)))))-- ( -21.50, z-score = -2.10, R) >droGri2.scaffold_15110 21248126 94 - 24565398 CCUUCCCCCCGCUAUCCG------CUGCAUCUACGCCUGACUGACACUUUUGUGUCGUGUCAUGCGUUUAAAUUGAGAG---AAAAAAUUGAACGUCC---CGUCA------ ..........((......------..))......((.((((((((((....)))))).)))).))((((((.((.....---...)).))))))....---.....------ ( -21.10, z-score = -2.58, R) >consensus AUGGCCCUCGGGGGCUUUA_____GU___UCCCCGCUUGAUUGACACUUUUGUGUCGCGUCAUGCGUUUAAAUUGUG_____AAAAACUCUCGCAUCCUCGCG_CA______ .....................................((((.(((((....)))))..)))).................................................. ( -6.43 = -6.44 + 0.01)


| Location | 15,748,152 – 15,748,245 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 66.17 |
| Shannon entropy | 0.61095 |
| G+C content | 0.47108 |
| Mean single sequence MFE | -23.34 |
| Consensus MFE | -6.33 |
| Energy contribution | -6.30 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3L 15748152 93 - 24543557 ---------------------UCCAUCAGAAUUCUUUGAAUGGCCCUCGGGGGCUUUAUGUUAGU---UCCCGGCUUGAUUGACACUUUUGUGUCUCGUCAUGCGUUUAAAUUGUGA-- ---------------------.((((((((....)))).))))....((((((((.......)))---)))))((.((((.(((((....)))))..)))).)).............-- ( -29.10, z-score = -2.78, R) >droSim1.chr3L 15088170 96 - 22553184 ---------------------UCCAUCAGAAUUCUUUGAAUGGCCCUCGGGGGCUUUAUGUUAGUUCCUCCCGGCUUGAUUGACACUUUUGUGUCUCGUCAUGCGUUUAAAUUGUGA-- ---------------------....(((.((((...(((((((((...(((((((.......)))))))...))).((((.(((((....)))))..))))..)))))))))).)))-- ( -27.40, z-score = -2.00, R) >droSec1.super_0 7858893 93 - 21120651 ---------------------UCCAUCAGAAUUCCUUGAAUGACCCUCGGGAGCUUUAUGUUAGU---UUCCGGCUUGAUUGACACUUUUGUGUCUCGUCAUGCGUUUAAAUUGUGA-- ---------------------....(((.((((...(((((......((((((((.......)))---)))))((.((((.(((((....)))))..)))).))))))))))).)))-- ( -26.60, z-score = -2.73, R) >droYak2.chr3L 18299873 88 - 24197627 ---------------------UCCAUCAGAAUUCCUUGAAUGGCCCUCGGGGGCU-----UUAGU---UCCCGGCUUGAUUGACACUUUUGUGUCUCGUCAUCCGUUUAAAUUGUGA-- ---------------------....(((.((((...(((((((...((((((((.-----...))---))))))..((((.(((((....)))))..)))).))))))))))).)))-- ( -28.80, z-score = -2.86, R) >droEre2.scaffold_4784 18003642 87 - 25762168 ---------------------UCCAUCAGAAUUCCUCAAAGGG-CCUCGGGCGCU-----UUAGU---UCCCGGCUUGAUUGACACUUUUGUGUCUCGUCAUCCGCUUAAAUUGUGA-- ---------------------....(((.((((....((((.(-((...))).))-----))...---...(((..((((.(((((....)))))..)))).)))....)))).)))-- ( -23.00, z-score = -1.12, R) >droAna3.scaffold_13337 11885444 89 + 23293914 ---------------------CGCAUCAGUGUCUCUUGAAUGGCCCUCUGGGGCUU----UUGGC---UCCUCGCUUGAUUGACACUUUUGUGUUGUGUCAUCCGUUUAAAUUGUGA-- ---------------------((((((((((....(..((.(((((....))))))----)..).---....))).))).((((((.........))))))...........)))).-- ( -22.80, z-score = -1.24, R) >dp4.chrXR_group6 13271046 88 + 13314419 ---------------------UCCAUCACAAUUCCUUGAAUGGCCCUCGGGGCCU-----UUGGC---CUUUCGCUUGAUUGACAAUUUUGUGUUGUCUCAUCUGUUUAAAUUAUGA-- ---------------------..............(((((((((((...))))).-----..(((---.....))).(((.((((((.....))))))..))).)))))).......-- ( -17.10, z-score = -0.03, R) >droPer1.super_12 1694097 88 + 2414086 ---------------------UCCAUCACAAUUCCUUGAAUGGCCCUCGGGGCCU-----UUGGC---CUUUCGCUUGAUUGACAAUUUUGUGUUGUCUCAUCUGUUUAAAUUAUGA-- ---------------------..............(((((((((((...))))).-----..(((---.....))).(((.((((((.....))))))..))).)))))).......-- ( -17.10, z-score = -0.03, R) >droMoj3.scaffold_6680 7860003 114 + 24764193 CUCCGCCUCCUCCCGCUUCUCCCGCUCCUCCUGCUGCUGAUCUACCCUUCGAUAC-----UCUGUGCAUCUUCGCCUGACUGACACUUUUGUGUUGUGUCAUGCGUUCAAAUUGAGAGA ....((........))(((((..........(((.((..(((........)))..-----...)))))....(((.((((((((((....)))))).)))).)))........))))). ( -21.10, z-score = -1.27, R) >droVir3.scaffold_13049 2939728 72 + 25233164 ----------------------------------------CCCCCCACACGGACC-----ACCUCUCCCCUUCGCCUGACUGACACUUUUGUGUUGUGUCAUGCGUUUAAAUUGAGA-- ----------------------------------------....((....))...-----..(((.......(((.((((((((((....)))))).)))).)))........))).-- ( -18.06, z-score = -3.31, R) >droGri2.scaffold_15110 21248146 91 - 24565398 ---------------------CCCCUCAACUCGCCGCU--CCUUCCCCCCGCUAU-----CCGCUGCAUCUACGCCUGACUGACACUUUUGUGUCGUGUCAUGCGUUUAAAUUGAGAGA ---------------------...(((((...((.((.--...............-----..)).))....((((.((((((((((....)))))).)))).)))).....)))))... ( -25.67, z-score = -4.64, R) >consensus _____________________UCCAUCAGAAUUCCUUGAAUGGCCCUCGGGGGCU_____UUAGU___UCCCCGCUUGAUUGACACUUUUGUGUCGUGUCAUGCGUUUAAAUUGUGA__ ............................................................................((((.(((((....)))))..)))).................. ( -6.33 = -6.30 + -0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:29:49 2011