| Sequence ID | dm3.chr3L |
|---|---|
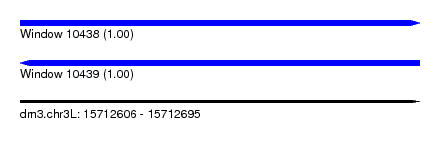
| Location | 15,712,606 – 15,712,695 |
| Length | 89 |
| Max. P | 0.999553 |

| Location | 15,712,606 – 15,712,695 |
|---|---|
| Length | 89 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.35 |
| Shannon entropy | 0.38640 |
| G+C content | 0.48607 |
| Mean single sequence MFE | -38.28 |
| Consensus MFE | -29.76 |
| Energy contribution | -30.04 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.96 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.01 |
| SVM RNA-class probability | 0.999553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
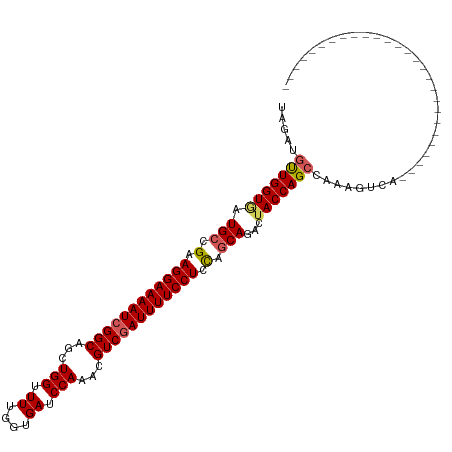
>dm3.chr3L 15712606 89 + 24543557 UAGAUAUUGGUGAUGCCGAAGGAAAAUCGGCAGCUGGUUUUGGUGAUCCAAACGUCGAUUUUCCUCCAGCAGACUACCAGCCAAAGUCA--------------------------- ..(((.(((((..(((.(.((((((((((((...(((.((....)).)))...)))))))))))).).)))........))))).))).--------------------------- ( -30.70, z-score = -3.14, R) >droEre2.scaffold_4784 17968797 116 + 25762168 UAGAUGCUGGUAAUGCCGCAGGAAAAUCGGCCGCUGGCUUUGGUGAUCCAACCGUCAAUUUUCCUCCGGCAGACCACCAGCCAAAUGCUGAUCCCUCCUGGCAAAUCCAUUCGUCA ..((((.(((...((((..((((..((((((...(((((..((((.((...(((............)))..)).)))))))))...))))))...))))))))...)))..)))). ( -39.00, z-score = -2.16, R) >droYak2.chr3L 18263309 116 + 24197627 UAGAUGCUGGUAAUGGCGAAGGAAAAUCGGCCGCUGGCUUUGGUGAUCCAACCGUCGAUUUUCCUUCGGCAGACUACCAGCCAAUCGAUGAUCCUUCAUGGAAAAUCCAUUCGUCA ..((((((((((.((.(((((((((((((((.(.(((.((....)).))).).))))))))))))))).))...)))))))..)))(((((......((((.....))))))))). ( -50.30, z-score = -5.24, R) >droSec1.super_0 7823685 89 + 21120651 UAGAUGUUGGUGAUGCCGUAGGAAAAUCGGCAGCUGGUUUUGGUGAUCCAAAUGUCGAUUUUCCUCUAGCAGACUACCAGCCAAAGUCA--------------------------- ..((((((((((.(((...(((((((((((((..(((.((....)).)))..)))))))))))))...)))...)))))))....))).--------------------------- ( -35.10, z-score = -4.46, R) >droSim1.chr3L 15051593 89 + 22553184 UAGAUGUUGGUGAUGCCGUAGGAAAAUCGGCAGCUGGUUUUGGUGAUCCAAAUGUCGAUUUUCCUCCAGCAGACUACCAGCCAAAGUCA--------------------------- ..((((((((((.(((.(.(((((((((((((..(((.((....)).)))..))))))))))))).).)))...)))))))....))).--------------------------- ( -36.30, z-score = -4.77, R) >consensus UAGAUGUUGGUGAUGCCGAAGGAAAAUCGGCAGCUGGUUUUGGUGAUCCAAACGUCGAUUUUCCUCCAGCAGACUACCAGCCAAAGUCA___________________________ .....(((((((.(((.(.((((((((((((...(((.((....)).)))...)))))))))))).).)))...)))))))................................... (-29.76 = -30.04 + 0.28)

| Location | 15,712,606 – 15,712,695 |
|---|---|
| Length | 89 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.35 |
| Shannon entropy | 0.38640 |
| G+C content | 0.48607 |
| Mean single sequence MFE | -37.74 |
| Consensus MFE | -26.44 |
| Energy contribution | -26.60 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.81 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15712606 89 - 24543557 ---------------------------UGACUUUGGCUGGUAGUCUGCUGGAGGAAAAUCGACGUUUGGAUCACCAAAACCAGCUGCCGAUUUUCCUUCGGCAUCACCAAUAUCUA ---------------------------.((..((((.(((.....((((((((((((((((.((.((((..........)))).)).)))))))))))))))))))))))..)).. ( -33.60, z-score = -4.31, R) >droEre2.scaffold_4784 17968797 116 - 25762168 UGACGAAUGGAUUUGCCAGGAGGGAUCAGCAUUUGGCUGGUGGUCUGCCGGAGGAAAAUUGACGGUUGGAUCACCAAAGCCAGCGGCCGAUUUUCCUGCGGCAUUACCAGCAUCUA ...((((((((((..((....))))))..))))))((((((((..(((((.((((((((((.(.(((((..........))))).).)))))))))).)))))))))))))..... ( -49.80, z-score = -3.51, R) >droYak2.chr3L 18263309 116 - 24197627 UGACGAAUGGAUUUUCCAUGAAGGAUCAUCGAUUGGCUGGUAGUCUGCCGAAGGAAAAUCGACGGUUGGAUCACCAAAGCCAGCGGCCGAUUUUCCUUCGCCAUUACCAGCAUCUA .....(((.(((..(((.....)))..))).))).(((((((((..((.((((((((((((.(.(((((..........))))).).)))))))))))))).)))))))))..... ( -48.10, z-score = -4.58, R) >droSec1.super_0 7823685 89 - 21120651 ---------------------------UGACUUUGGCUGGUAGUCUGCUAGAGGAAAAUCGACAUUUGGAUCACCAAAACCAGCUGCCGAUUUUCCUACGGCAUCACCAACAUCUA ---------------------------.((..((((.(((.....((((..((((((((((.((.((((..........)))).)).))))))))))..)))))))))))..)).. ( -27.60, z-score = -3.21, R) >droSim1.chr3L 15051593 89 - 22553184 ---------------------------UGACUUUGGCUGGUAGUCUGCUGGAGGAAAAUCGACAUUUGGAUCACCAAAACCAGCUGCCGAUUUUCCUACGGCAUCACCAACAUCUA ---------------------------.((..((((.(((.....(((((.((((((((((.((.((((..........)))).)).)))))))))).))))))))))))..)).. ( -29.60, z-score = -3.44, R) >consensus ___________________________UGACUUUGGCUGGUAGUCUGCUGGAGGAAAAUCGACGUUUGGAUCACCAAAACCAGCUGCCGAUUUUCCUACGGCAUCACCAACAUCUA ...................................(.((((....(((((.((((((((((.((.((((..........)))).)).)))))))))).)))))..)))).)..... (-26.44 = -26.60 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:29:38 2011