| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,677,318 – 15,677,448 |
| Length | 130 |
| Max. P | 0.646867 |

| Location | 15,677,318 – 15,677,448 |
|---|---|
| Length | 130 |
| Sequences | 11 |
| Columns | 140 |
| Reading direction | forward |
| Mean pairwise identity | 66.44 |
| Shannon entropy | 0.70944 |
| G+C content | 0.54599 |
| Mean single sequence MFE | -44.62 |
| Consensus MFE | -16.17 |
| Energy contribution | -15.97 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
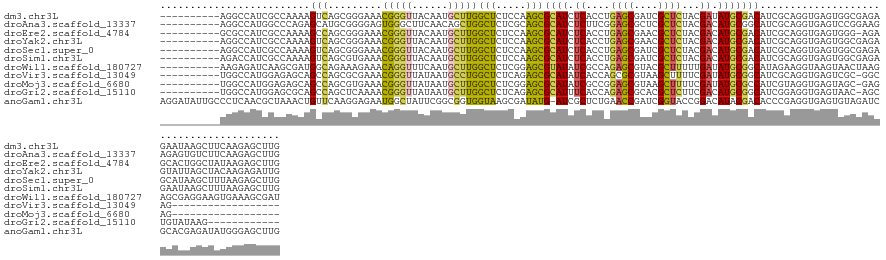
>dm3.chr3L 15677318 130 + 24543557 ----------AGGCCAUCGCCAAAAGUCAGCGGGAAACGGGUUACAAUGCUUGGCUCUCCAAGCGCAUCUCACCUGAGCGAUCGCUCUACGAUAUGCGACAUCGCAGGUGAGUGGCGAGAGAAUAAGCUUCAAGAGCUUG ----------......((((((...((.(.(.(....).).).))...((((((....))))))....(((((((((((....))))........(((....)))))))))))))))).....(((((((...))))))) ( -47.40, z-score = -1.77, R) >droAna3.scaffold_13337 11817873 130 - 23293914 ----------AGGCCAUGGCCCAGAGCAUGCGGGGAGUGGGCUUCAACAGCUGGCUCUCGCAGCGCAUCUCUUCGGAGCGCUCGCUCUACGACAUGCGGCAUCGCAGGUGAGUCCGGAAGAGAGUGUCUUCAAGAGCUUG ----------.(((....)))..((((.((((((.(((.((((.....)))).)))))))))((((.((((((((((((....)))))..(((.((((....)))).....)))..)))))))))))........)))). ( -53.90, z-score = -0.34, R) >droEre2.scaffold_4784 17934968 129 + 25762168 ----------GCGCCAUCGCCAAAAGCCAGCGGGAAACGGGUUACAAUGCUUGGCUCUCCAAGCGCAUCUCACCUGAGCGAACGCUCUACGACAUGCGACAUCGCAGGUGAGUGGG-AGAGCACUGGCUAUAAGAGCUUG ----------((((....))....((((((.......(((((......)))))(((((((.....((.(((((((((((....))))........(((....))))))))))))))-))))).))))))......))... ( -48.70, z-score = -1.77, R) >droYak2.chr3L 18227892 130 + 24197627 ----------AGGCCAUCGCCAAAAGUCAGCGGGAAACGGGUUACAAUGCUUGGCUCUCCAAGCGCAUCUCACCUGAGCGAACGCUCUACGACAUGCGACAUCGCAGGUGAGUGGCGAGAGUAUUAGCUACAAGAGAUUG ----------.(((....)))...((((..(.(....).)........(((..((((((...((.((.(((((((((((....))))........(((....))))))))))))))))))))...))).......)))). ( -44.90, z-score = -1.54, R) >droSec1.super_0 7788590 130 + 21120651 ----------AGGCCAUCGCCAAAAGUCAGCGGGAAACGGGUUACAAUGCUUGGCUCUCCAAGCGCAUCUCACCUGAGCGAUCGCUCUACGACAUGCGACAUCGCAGGUGAGUGGCGAGAGCAUAAGCUUUAAGAGCUUG ----------..((..((((((...((.(.(.(....).).).))...((((((....))))))....(((((((((((....))))........(((....))))))))))))))))..)).(((((((...))))))) ( -51.20, z-score = -2.68, R) >droSim1.chr3L 15016446 130 + 22553184 ----------AGACCAUCGCCAAAAGUCAGCGUGAAACGGGUUACAAUGCUUGGCUCUCCAAGCGCAUCUCACCUGAGCGAUCGCUCUACGACAUGCGACAUCGCAGGUGAGUGGCGAGAGAAUAAGCUUUAAGAGCUUG ----------...(..((((((.......((((((......))))...((((((....))))))))..(((((((((((....))))........(((....))))))))))))))))..)..(((((((...))))))) ( -47.60, z-score = -2.42, R) >droWil1.scaffold_180727 1076097 130 - 2741493 ----------AAGAGAUCAAGCGAUGGCAGAAAGAAACAGGUUUCAAUGCUUGGCUCUCGGAGCGUAUAUCGCCAGAGCGUACGCUUUUUGAUAUGCGGCAUAGAAGGUAAGUAACUAAGAGCGAGGAAGUGAAAGCGAU ----------.....(((..((....))...........(.(((((...(((.(((((..(..(((((((((..(((((....))))).)))))))))((.......))......)..))))).)))...))))).)))) ( -36.00, z-score = -1.06, R) >droVir3.scaffold_13049 2862366 111 - 25233164 ----------UGGCCAUGGAGAGCAGCCAGCGCGAAACGGGUUAUAAUGCCUGGCUCUCAGAGCGCAUAUCACCAGCGCGUAAGCUUUUCGAUAUGCGGCAUCGCAGGUGAGUCGC-GGCAG------------------ ----------..(((...((((((.((....))....(((((......)))))))))))...(((....(((((...((....)).........((((....)))))))))..)))-)))..------------------ ( -44.10, z-score = -1.31, R) >droMoj3.scaffold_6680 7778975 111 - 24764193 ----------UGGCCAUGGAGAGCAGCCAGCGUGAAACGGGUUAUAAUGCUUGGCUCUCGGAGCGCAUAUCGCCGGAGCGUAAGCUUUUCGAUAUGCGCCAUCGUAGGUGAGUAGC-GAGAG------------------ ----------(.(((((((.(((.(((((((((.............)))).)))))))).(.((((((((((..(((((....))))).))))))))))).)))).))).).....-.....------------------ ( -43.82, z-score = -1.94, R) >droGri2.scaffold_15110 21172520 117 + 24565398 ----------UGGCCAUGGAGCGCAGCCAGCUCAAAACGGGUUAUAAUGCUUGGCUCUCAGAGCGCAUUUCACCAGAGCGCACGCUCUUCGACAUGCGGCAUCGGAGGUGAGUAAC-AGCUGUAUAAG------------ ----------..((......))(((((.(((((.....)))))....((((((.((((..((((((((.((...(((((....)))))..)).)))).)).)))))).))))))..-.))))).....------------ ( -35.70, z-score = 0.47, R) >anoGam1.chr3L 35191161 139 + 41284009 AGGAUAUUGCCCUCAACGCUAAACUGUUCAAGGAGAAUGGCUAUUCGGCGGUGGUAAGCGAUAUG-AUCGCUCUGAACCGAUCGGUACCGGACAUACGACACCCGAGGUGAGUGUAGAUCGCACGAGAUAUGGGAGCUUG ........((.((((.((((...((((((.....))))))....((((..((.(((.(((((...-)))))((((.(((....)))..))))..))).))..)))))))).((((.....))))......)))).))... ( -37.50, z-score = 0.91, R) >consensus __________AGGCCAUCGCCAAAAGCCAGCGGGAAACGGGUUACAAUGCUUGGCUCUCCAAGCGCAUCUCACCUGAGCGAACGCUCUACGACAUGCGACAUCGCAGGUGAGUGGCGAGAGGAUAAGCUUUAAGAGCUUG .........................(((.........(((((......)))))(((.....)))((((.((....((((....))))...)).)))))))........................................ (-16.17 = -15.97 + -0.20)
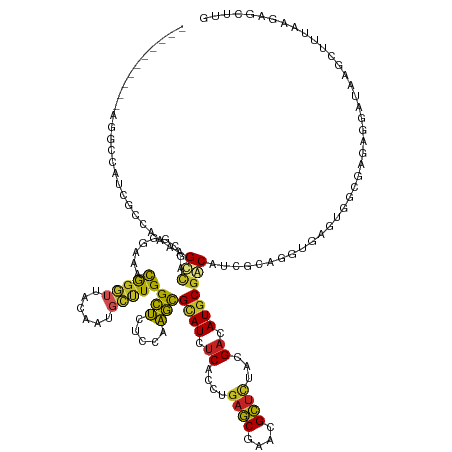

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:29:32 2011