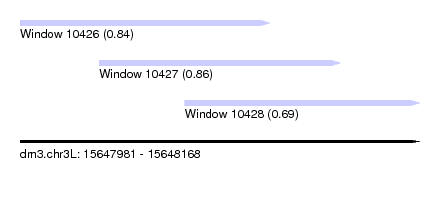
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,647,981 – 15,648,168 |
| Length | 187 |
| Max. P | 0.856397 |

| Location | 15,647,981 – 15,648,098 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.12 |
| Shannon entropy | 0.35363 |
| G+C content | 0.52924 |
| Mean single sequence MFE | -43.52 |
| Consensus MFE | -26.95 |
| Energy contribution | -27.75 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.840702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15647981 117 + 24543557 CCUCCGGGAUUCCAACCUGAAGGUUUUGUCGAAC---AUUCGUCCGGUUUCUUGGUAGUCGUUGAAAAGCUUGGGAUUAUAGGACCGCCAGGAUUCCAUCCAGACGGGUGAGAGCUACUA (((.((((.......)))).)))..........(---(((((((.((..((((((..((((((....)))(((......))))))..))))))......)).)))))))).......... ( -33.80, z-score = 0.42, R) >droSim1.chr3L 14987350 117 + 22553184 CCGUCGGGAUUCCAACCUGACGGUUUUCUCGAGC---AUUCGUCUGGUUUCUUGGUAGUCGUUGAACCGCUUGGGAUUAUAGGACCGCCAGGAUUCCAUCCAGACGGGUGAGAGCUACUA ((((((((.......))))))))....(((...(---((((((((((..((((((..(((.((((.((.....)).))).).)))..))))))......))))))))))).)))...... ( -49.40, z-score = -3.49, R) >droSec1.super_0 7760214 117 + 21120651 CCGCCGGGAUUCCAACCUGACGGUUUUCUCGAGC---AUUCGUCUGGUUUCUUGGUAGUCGUUGAACCGCUUGGUAUUAUAGGACCGCCAGGAUUCCAUCCAGACGGGUGAGAGCUACUA (((.((((.......)))).)))....(((...(---((((((((((..((((((..(((.(((((((....))).))).).)))..))))))......))))))))))).)))...... ( -44.30, z-score = -2.40, R) >droYak2.chr3L 18197604 117 + 24197627 CCGCCGGGAUUCCAGCCCGAAGGUUCUUGCGAACCACAUCCACAUGGUUUCUUGGUGG---UUGAGACUAUUGGGAUUAUAGGACCGCCAGGAUUCCAUCCUGGUGGGCGAGUUUCACCA (((((((((..(((....((.(((((....)))))...))....)))..)))))))))---.(((((((.(((......)))..((((((((((...))))))))))...)))))))... ( -46.30, z-score = -2.10, R) >droEre2.scaffold_4784 17906041 114 + 25762168 CCGCCGGGAUUACAACCUGAAUUUGGCGAGCCAC---AUUCGUCUGGUUUCUUGGUAGUCCUUGGAGAGAUUGGGAUUAACGGACCGCCAGGAUUCCAACCAGAUGGGUGAGUGCGA--- .(((((((.(((.....))).))))))).(((.(---((((((((((((((((((..((((...((..........))...))))..))))))....))))))))))))).).))..--- ( -43.80, z-score = -2.30, R) >consensus CCGCCGGGAUUCCAACCUGAAGGUUUUCUCGAAC___AUUCGUCUGGUUUCUUGGUAGUCGUUGAAACGCUUGGGAUUAUAGGACCGCCAGGAUUCCAUCCAGACGGGUGAGAGCUACUA (((.((((.......)))).)))..............((((((((((..(((((((.(((......................))).)))))))......))))))))))........... (-26.95 = -27.75 + 0.80)

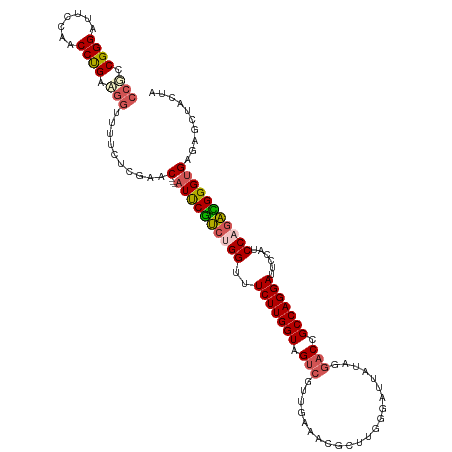
| Location | 15,648,018 – 15,648,131 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.10 |
| Shannon entropy | 0.52821 |
| G+C content | 0.52280 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -19.89 |
| Energy contribution | -20.57 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.856397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15648018 113 + 24543557 CGUCCGGUUUCUUGGUAGUCGUUGAAAAGCUUGGGAUUAUAGGACCGCCAGGAUUCCAUCCAGACGGGUGAGAGCUACUACAUCCGCAUGGUUUUCGGGUGGUUUUCUCUGGA------- .....((..((((((..((((((....)))(((......))))))..))))))..)).((((((.....((((((((((....((....))......))))))))))))))))------- ( -37.60, z-score = -0.88, R) >droSim1.chr3L 14987387 88 + 22553184 CGUCUGGUUUCUUGGUAGUCGUUGAACCGCUUGGGAUUAUAGGACCGCCAGGAUUCCAUCCAGACGGGUGAGAGCUACUACAACCGCA-------------------------------- (((((((..((((((..(((.((((.((.....)).))).).)))..))))))......)))))))(((....)))............-------------------------------- ( -30.70, z-score = -1.63, R) >droSec1.super_0 7760251 88 + 21120651 CGUCUGGUUUCUUGGUAGUCGUUGAACCGCUUGGUAUUAUAGGACCGCCAGGAUUCCAUCCAGACGGGUGAGAGCUACUACAACCGCA-------------------------------- (((((((..((((((..(((.(((((((....))).))).).)))..))))))......)))))))(((....)))............-------------------------------- ( -30.60, z-score = -1.93, R) >droYak2.chr3L 18197644 111 + 24197627 CACAUGGUUUCUUGGUGGU---UGAGACUAUUGGGAUUAUAGGACCGCCAGGAUUCCAUCCUGGUGGGCGAGUUUCACCACAUCCACAUGGCUUUCGGGUGGUGGUCGUUGAGA------ .........(((..(((((---....)))))..)))........((((((((((...))))))))))((((.(.(((((....((....))......))))).).)))).....------ ( -38.60, z-score = -0.78, R) >droEre2.scaffold_4784 17906078 117 + 25762168 CGUCUGGUUUCUUGGUAGUCCUUGGAGAGAUUGGGAUUAACGGACCGCCAGGAUUCCAACCAGAUGGGUGAG---UGCGACAUCCGCAUGGUUUUUCGGUGGUUGUCGAAGCUGUUGUCA (((((((((((((((..((((...((..........))...))))..))))))....)))))))))(((...---..(((((.((((.(((....))))))).)))))..)))....... ( -42.10, z-score = -1.60, R) >consensus CGUCUGGUUUCUUGGUAGUCGUUGAAACGCUUGGGAUUAUAGGACCGCCAGGAUUCCAUCCAGACGGGUGAGAGCUACUACAUCCGCAUGG_UUU__GGUGGU__UC___G_________ .((((((..(((((((.(((......................))).)))))))......))))))(((((..........)))))................................... (-19.89 = -20.57 + 0.68)



| Location | 15,648,058 – 15,648,168 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 60.97 |
| Shannon entropy | 0.54668 |
| G+C content | 0.55180 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -20.98 |
| Energy contribution | -20.99 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.686536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15648058 110 + 24543557 AGGACCGCCAGGAUUCCAUCCAGACGGGUGAGAGCUACUACAUCCGCAUGGUUUUCGGGUGGUUUUCU------CUG-GAGGAGGAGUGCUAUUUGGUGGUUCGGUGUUCUUAGUGC--- .((((((((((.(((((.(((...((((.((((((((((....((....))......)))))))))))------)))-..)))))))).)....)))))))))..............--- ( -41.40, z-score = -1.90, R) >droYak2.chr3L 18197681 104 + 24197627 AGGACCGCCAGGAUUCCAUCCUGGUGGGCGAGUUUCACCACAUCCACAUGGCUUUCGGGUGGUGGUCG------UUGAGACGAUAGGGAUUAUAGGACCGCCUGGAUUCC---------- ....((((((((((...))))))))))....((((((..((..((((...(((....))).))))..)------)))))))....((((((.((((....))))))))))---------- ( -38.40, z-score = -0.74, R) >droEre2.scaffold_4784 17906118 116 + 25762168 CGGACCGCCAGGAUUCCAACCAGAUGGGUGAG---UGCGACAUCCGCAUGGUUUUUCGGUGGUUGUCGAAGCUGUUGUCACUGGAGGAGGUCUAUG-UGGCUUAGUCCUUCGAGUAGUGC ..(((((((..((..((.(((.....)))..(---((((.....)))))))..))..))))))).(((((((((..(((((((((.....)))).)-)))).)))..))))))....... ( -39.90, z-score = -0.65, R) >consensus AGGACCGCCAGGAUUCCAUCCAGAUGGGUGAG___UACCACAUCCGCAUGGUUUUCGGGUGGUUGUCG______UUG_GACGAGAGGAGCUAUAUG_UGGCUUGGUCUUC__AGU_____ ..(((((((.(((..((((...(.((((((..........)))))))))))..))).))))))).....................(((((................)))))......... (-20.98 = -20.99 + 0.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:29:29 2011