
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,625,277 – 15,625,394 |
| Length | 117 |
| Max. P | 0.625359 |

| Location | 15,625,277 – 15,625,394 |
|---|---|
| Length | 117 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.37 |
| Shannon entropy | 0.34426 |
| G+C content | 0.53277 |
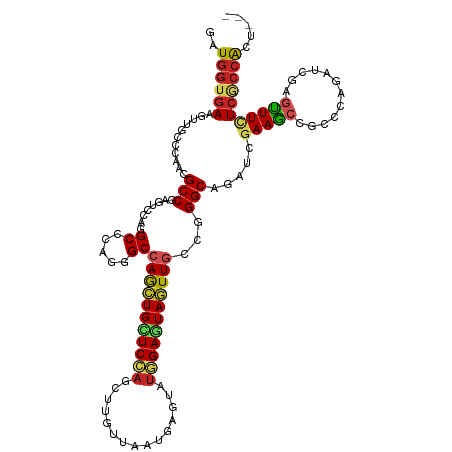
| Mean single sequence MFE | -37.28 |
| Consensus MFE | -22.34 |
| Energy contribution | -22.37 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.625359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
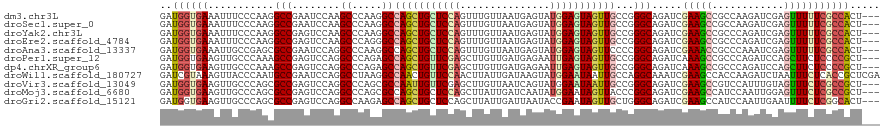
>dm3.chr3L 15625277 117 + 24543557 GAUGGUGAAAUUUCCCAAGGCCGAAUCCAAGCCCAAGGCCAGCUGCUCCAGUUUGUUAAUGAGUAUGGAGUAGUUGCCGGGCAGAUCGAAGCCGCCAAGAUCGAGUUUUUCGCCACU--- ..((((((((.(((................((((..(((.((((((((((...............))))))))))))))))).((((...........)))))))..))))))))..--- ( -40.86, z-score = -2.87, R) >droSec1.super_0 7737870 117 + 21120651 GAUGGUGAAAUUUCCCAAGGCCGAAUCCAAGCCCAAGGCCAGCUGCUCCAGUUUGUUAAUGAGUAUGGAGUAGUUGCCGGGCAGAUCGAAGCCGCCAAGAUCGAGUUUUUCGCCACU--- ..((((((((.(((................((((..(((.((((((((((...............))))))))))))))))).((((...........)))))))..))))))))..--- ( -40.86, z-score = -2.87, R) >droYak2.chr3L 18173414 117 + 24197627 GAUGGUGAAAUUUCCCAAGGCCGAGUCCAAGCCCAAGGCCAGCUGCUCCAGUUUGUUAAUGAGUAUGGAGUAGUUGCCGGGCAGAUCGAAGCCGCCCAGAUCGAGUUUUUCGCCACU--- ..((((((((.(((....(((.(..((...((((..(((.((((((((((...............))))))))))))))))).....))..).)))......)))..))))))))..--- ( -41.46, z-score = -2.91, R) >droEre2.scaffold_4784 17883177 117 + 25762168 GAUGGUGAAAUUUCCCAAGGCCGAGUCCAAGCCCAGGGCCAGCUGCUCCAGUUUGUUAAUGAGUAUGGAGUAGUUGCCGGGCAGAUCGAAGCCGCCCAGAUCGAGUUUUUCGCCACU--- ..((((((((.(((....(((.(..((...((((..(((.((((((((((...............))))))))))))))))).....))..).)))......)))..))))))))..--- ( -41.46, z-score = -2.41, R) >droAna3.scaffold_13337 11760609 117 - 23293914 GAUGGUGAAAUUGCCGAGCGCCGAAUCCAGGCCCAAGGCCAGCUGCUCCAGUUUGUUAAUGAGUAUGGAGUAGUUCCCCGGCAGAUCGAAACCGCCCAAAUCGAGUUUUUCGCCACU--- ..((((((((..(((((..((((......((((...))))((((((((((...............))))))))))...)))).(..((....))..)...))).)).))))))))..--- ( -39.86, z-score = -2.95, R) >droPer1.super_12 1589890 117 - 2414086 GAUGGUGAAGUUGCCCAAAGCCGAGUCCAGGCCCAGAGCCAGCUGUUCGAGCUUGUUGAUGAGAAUUGAGUAGUUGCCGGGCAGAUCAAAGCCGCCCAGAUCCAGCUUCUCCCCGCU--- ...((.((((((((((...(((.......))).....(.(((((((((((.(((......)))..))))))))))).))))).((((...........)))).)))))).)).....--- ( -34.10, z-score = -0.48, R) >dp4.chrXR_group6 13174695 117 - 13314419 GAUGGUGAAGUUGCCCAAAGCCGAGUCCAGGCCCAGAGCCAGCUGUUCGAGCUUGUUGAUGAGAAUUGAGUAGUUGCCGGGCAGAUCAAAGCCGCCCAGAUCCAGCUUCUCCCCGCU--- ...((.((((((((((...(((.......))).....(.(((((((((((.(((......)))..))))))))))).))))).((((...........)))).)))))).)).....--- ( -34.10, z-score = -0.48, R) >droWil1.scaffold_180727 1018098 120 - 2741493 GAUCGUAAAGUUACCCAAUGCCGAAUCCAGGCCUAAGGCCAACUGUUCCAACUUAUUGAUAAGUAUGGAAUAAUUGCCAGGCAAAUCGAAGCCACCAAGAUCUAAUUUCUCACCGCUCGA ((((...............(((.......)))....(((.((.((((((((((((....))))).))))))).))))).(((........))).....)))).................. ( -27.80, z-score = -1.72, R) >droVir3.scaffold_13049 2802301 117 - 25233164 GAUGGUGAAGUUGCCCAGCGCCGAGUCCAGGCCCAGCGCCAAUUGUUCGAGCUUGUUAAUCAGUAUGGAAUAAUUGCCGGGCAGAUCGAAGCCGUCCAUUUGUAGUUUCUCGCCGCU--- ...(((((..((((((.((((.(.((....)).).))))(((((((((.(..(((.....)))..).)))))))))..))))))...(((((..(......)..))))))))))...--- ( -37.60, z-score = -1.34, R) >droMoj3.scaffold_6680 7730707 117 - 24764193 GAUGGUGAAGUUGCCCAGCGCCGAGUCCAGGCCCAGCGCCAGCUGCUCCAGCUUAUUGAUCAAUAUGGAAUAGUUACCCGGCAGAUCGAAGCCAUCCAAUUGGAGUUUCUCGCCGCU--- ...(((((..(((((..((((.(.((....)).).)))).(((((.((((...((((....)))))))).)))))....)))))...(((((..(((....)))))))))))))...--- ( -35.60, z-score = -0.53, R) >droGri2.scaffold_15121 147542 117 - 279557 GAUGGUGAAGUUGCCCAGCGCCGAGUCCAGGCCAAGAGCCAGCUGCUCCAGCUUAUUGAUUAAUACCGAAUAGUUGCUGGGCAGAUCGAAGCCAUCCAAUUGAAUUUUCUCGGCACU--- ((((((....((((((((((.(...((..(((.....)))(((((...)))))..............))...).))))))))))......)))))).....................--- ( -36.40, z-score = -1.13, R) >consensus GAUGGUGAAGUUGCCCAACGCCGAGUCCAGGCCCAGGGCCAGCUGCUCCAGCUUGUUAAUGAGUAUGGAGUAGUUGCCGGGCAGAUCGAAGCCGCCCAGAUCGAGUUUCUCGCCACU___ ..((((((...........(((........((.....))(((((((((((...............)))))))))))...))).....(((((............)))))))))))..... (-22.34 = -22.37 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:29:22 2011