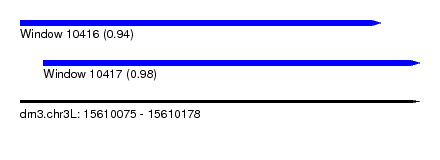
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,610,075 – 15,610,178 |
| Length | 103 |
| Max. P | 0.979516 |

| Location | 15,610,075 – 15,610,168 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 73.81 |
| Shannon entropy | 0.47098 |
| G+C content | 0.55068 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -12.70 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938029 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
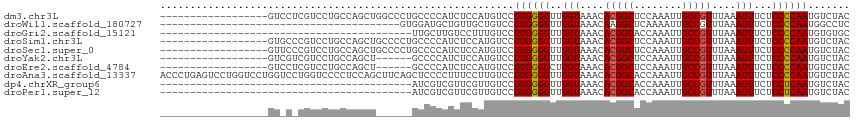
>dm3.chr3L 15610075 93 + 24543557 ----------------------GUAGACGUCCUCGUCCUG-CCAGCUGGCCCUGCCCCAUCUCCAUGUCCUUGGGGUUGGUAAACACGGCUCCAAAUUGCCGUUUAAAUUUCUCCC ----------------------...((((....))))..(-((....)))((.((((((............)))))).)).....(((((........)))))............. ( -27.20, z-score = -2.31, R) >droWil1.scaffold_180727 1001637 71 - 2741493 ---------------------------------------------GUGGAUGUGGAUGCUGUUGCUGUCCUUGGGGUUGGUAAACAAGGCUCAAAAUUGCCGUUUAAAUUUCUCCC ---------------------------------------------........((((((....)).))))..((((....(((((..(((........)))))))).....)))). ( -17.20, z-score = -0.54, R) >droSim1.chr3L 14949508 93 + 22553184 ----------------------GGAGCCGUGCCCGUCCUG-CCAGCUGCCCCUGCCCCAUCUCCAUGUCCUUGGGGUUGGUAAACACGGCUCCAAAUUGCCGUUUAAAUUUCUCCC ----------------------(((((((((...((....-...))((((...((((((............)))))).))))..)))))))))....................... ( -30.90, z-score = -2.79, R) >droSec1.super_0 7722749 93 + 21120651 ----------------------GGAGCCGUUCCCGUCCUG-CCAGCUGCCCCUGCCCCAUCUCCAUGUCCUUGGGGUUGGUAAACACGGCUCCAAAUUGCCGUUUAAAUUUCUCCC ----------------------((((((((....((..((-(((...(((((.....(((....))).....)))))))))).))))))))))....................... ( -28.70, z-score = -2.55, R) >droYak2.chr3L 18158140 87 + 24197627 ----------------------GAUGCCGUCGUCGUCCUG-CCAGCUG------CCCCAUCUCCAUGUCCUUGGGGUUGGUAAACACGGCUCCAAAUUGCCGUUUAAAUUUCUCCC ----------------------((((....))))((..((-(((...(------(((((............))))))))))).))(((((........)))))............. ( -24.00, z-score = -2.03, R) >droEre2.scaffold_4784 17868210 87 + 25762168 ----------------------GAUGCCGUCCUCGUCCUG-CCAGCUG------CCCCAUCUCCAUGUCCUUGGGGCUGGUAAACACGGCUCCAAAUUGCCGUUUAAAUUUCUCCC ----------------------((((.......)))).((-(((...(------(((((............)))))))))))...(((((........)))))............. ( -23.60, z-score = -2.01, R) >droAna3.scaffold_13337 11744984 114 - 23293914 GUUGCCUUUACCCUGAGUCCUGGUCCUGGUCCUGGUCCCCUCCAGCUUCAGC--UCCCCUUUCCUUGUCCUUGGGGUUGGUAAACACGGCACCAAAUUGCCGUUUAAAUUUCUCCC .(((((...((((..((..(.((..((((..((((......))))..)))).--.)).........)..))..)))).)))))..((((((......))))))............. ( -36.50, z-score = -3.55, R) >consensus ______________________GAAGCCGUCCUCGUCCUG_CCAGCUGCCCCUGCCCCAUCUCCAUGUCCUUGGGGUUGGUAAACACGGCUCCAAAUUGCCGUUUAAAUUUCUCCC ........................................................(((.(((((......))))).))).....(((((........)))))............. (-12.70 = -13.50 + 0.80)
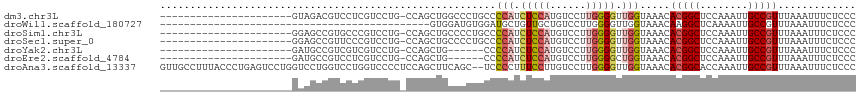
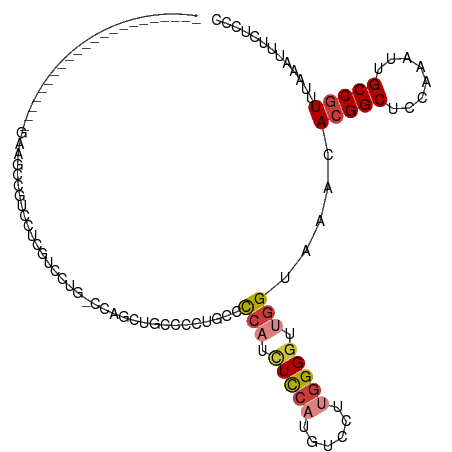
| Location | 15,610,081 – 15,610,178 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.63 |
| Shannon entropy | 0.51466 |
| G+C content | 0.51044 |
| Mean single sequence MFE | -21.43 |
| Consensus MFE | -16.25 |
| Energy contribution | -16.19 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979516 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 15610081 97 + 24543557 ------------------GUCCUCGUCCUGCCAGCUGGCCCUGCCCCAUCUCCAUGUCCUUGGGGUUGGUAAACACGGCUCCAAAUUGCCGUUUAAAUUUCUCCCCAAUGUCUAC ------------------...........(((....)))((.((((((............)))))).)).....(((((........)))))....................... ( -23.30, z-score = -1.43, R) >droWil1.scaffold_180727 1001643 75 - 2741493 ----------------------------------------GUGGAUGCUGUUGCUGUCCUUGGGGUUGGUAAACAAGGCUCAAAAUUGCCGUUUAAAUUUCUCCCCAAUGGCCUC ----------------------------------------..((((((....)).))))((((((....(((((..(((........)))))))).......))))))....... ( -21.40, z-score = -1.28, R) >droGri2.scaffold_15121 124759 73 - 279557 ------------------------------------------UUGCUUGUCCUUUGUCCUUGGGGUUGGUAAACACGGCACCAAAUUGCCGUUUAAAUUUCUCCCCAAUGUGUGC ------------------------------------------.................((((((..(((....((((((......))))))....)))...))))))....... ( -17.90, z-score = -0.84, R) >droSim1.chr3L 14949514 97 + 22553184 ------------------GUGCCCGUCCUGCCAGCUGCCCCUGCCCCAUCUCCAUGUCCUUGGGGUUGGUAAACACGGCUCCAAAUUGCCGUUUAAAUUUCUCCCCAAUGUCUAC ------------------(((..(((.........((((...((((((............)))))).))))...(((((........)))))...............)))..))) ( -21.70, z-score = -0.84, R) >droSec1.super_0 7722755 97 + 21120651 ------------------GUUCCCGUCCUGCCAGCUGCCCCUGCCCCAUCUCCAUGUCCUUGGGGUUGGUAAACACGGCUCCAAAUUGCCGUUUAAAUUUCUCCCCAAUGUCUAC ------------------......((..(((((...(((((.....(((....))).....)))))))))).))(((((........)))))....................... ( -21.60, z-score = -1.46, R) >droYak2.chr3L 18158146 91 + 24197627 ------------------GUCGUCGUCCUGCCAGCU------GCCCCAUCUCCAUGUCCUUGGGGUUGGUAAACACGGCUCCAAAUUGCCGUUUAAAUUUCUCCCCAAUGUCUAC ------------------......((..(((((...------((((((............))))))))))).))(((((........)))))....................... ( -22.20, z-score = -2.01, R) >droEre2.scaffold_4784 17868216 91 + 25762168 ------------------GUCCUCGUCCUGCCAGCU------GCCCCAUCUCCAUGUCCUUGGGGCUGGUAAACACGGCUCCAAAUUGCCGUUUAAAUUUCUCCCCAAUGUCUAC ------------------......((..(((((...------((((((............))))))))))).))(((((........)))))....................... ( -23.60, z-score = -2.73, R) >droAna3.scaffold_13337 11744993 115 - 23293914 ACCCUGAGUCCUGGUCCUGGUCCUGGUCCCCUCCAGCUUCAGCUCCCCUUUCCUUGUCCUUGGGGUUGGUAAACACGGCACCAAAUUGCCGUUUAAAUUUCUCCCCAAUGUCUAC .....(((....((..((((..((((......))))..))))..))...)))...(.(.((((((..(((....((((((......))))))....)))...)))))).).)... ( -31.20, z-score = -1.65, R) >dp4.chrXR_group6 13162049 73 - 13314419 ------------------------------------------AUCGUCGUUCGUUGUCCUUGGGGUUGGUAAACACGGCACCAAAUUGCCGUUUAAAUUUCUCCUCAAUGUCUAC ------------------------------------------..........((.(.(.((((((..(((....((((((......))))))....)))...)))))).).).)) ( -15.70, z-score = -1.19, R) >droPer1.super_12 1577205 73 - 2414086 ------------------------------------------AUCGUCGUUCGUUGUCCUUGGGGUUGGUAAACACGGCACCAAAUUGCCGUUUAAAUUUCUCCUCAAUGUCUAC ------------------------------------------..........((.(.(.((((((..(((....((((((......))))))....)))...)))))).).).)) ( -15.70, z-score = -1.19, R) >consensus __________________GU_C_CGUCCUGCCAGCU______GCCCCAUCUCCAUGUCCUUGGGGUUGGUAAACACGGCUCCAAAUUGCCGUUUAAAUUUCUCCCCAAUGUCUAC ...........................................................((((((..(((....(((((........)))))....)))...))))))....... (-16.25 = -16.19 + -0.06)
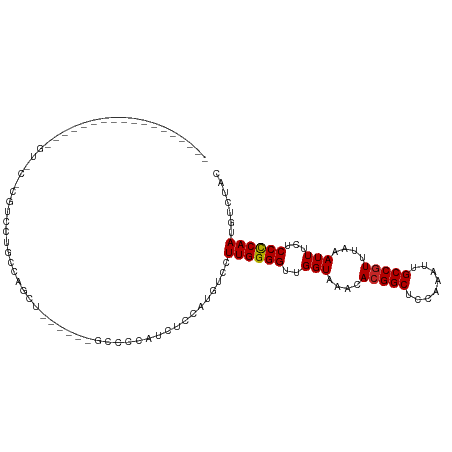
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:29:20 2011