| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,588,802 – 15,588,859 |
| Length | 57 |
| Max. P | 0.901862 |

| Location | 15,588,802 – 15,588,859 |
|---|---|
| Length | 57 |
| Sequences | 5 |
| Columns | 61 |
| Reading direction | forward |
| Mean pairwise identity | 68.07 |
| Shannon entropy | 0.56540 |
| G+C content | 0.27158 |
| Mean single sequence MFE | -10.16 |
| Consensus MFE | -5.41 |
| Energy contribution | -4.85 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
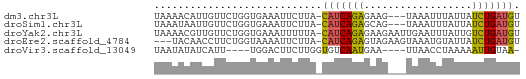
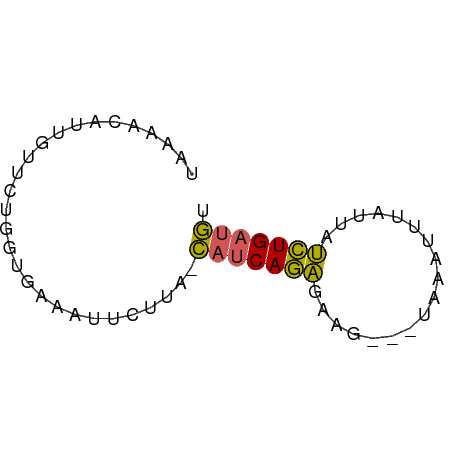
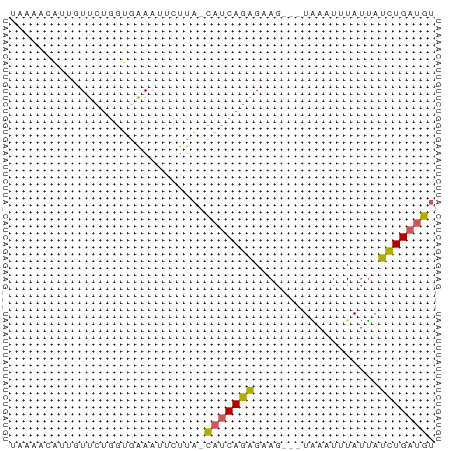
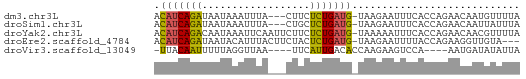
>dm3.chr3L 15588802 57 + 24543557 UAAAACAUUGUUCUGGUGAAAUUCUUA-CAUCAGAGAAG---UAAAUUUAUUAUCUGAUGU .....((((.....))))........(-(((((((..((---((....)))).)))))))) ( -10.30, z-score = -1.31, R) >droSim1.chr3L 14928751 57 + 22553184 UAAAUAAUUGUUCUGGUGAAAUUCUUA-CAUCAGAGCAG---UAAAUUUAUUAUCUGAUGU ......((((((((((((.........-)))))))))))---).................. ( -14.00, z-score = -3.23, R) >droYak2.chr3L 18136565 60 + 24197627 UAAAACGUUGUUCUGGUGAAAUUUUUA-CAUCAGAGAAGAAUUGAAUUUAUUGUCUGAUGU ..........(((....)))......(-(((((((.((.((......)).)).)))))))) ( -10.50, z-score = -0.88, R) >droEre2.scaffold_4784 17847193 57 + 25762168 ---UACAACCUUCUGGUAAAAUUCUUA-CAUCAGAGUAGAAGUAAAUGUAUUAUCUGAUGU ---....(((....))).........(-((((((((((.(......).)))..)))))))) ( -10.70, z-score = -1.41, R) >droVir3.scaffold_13049 2766615 52 - 25233164 UAAUAUAUCAUU----UGGACUUCUUGGUGUCAAUGAA----UUAACCUAAAAAUUGUAA- .......(((((----.(.(((....))).).))))).----..................- ( -5.30, z-score = -0.06, R) >consensus UAAAACAUUGUUCUGGUGAAAUUCUUA_CAUCAGAGAAG___UAAAUUUAUUAUCUGAUGU ............................(((((((..................))))))). ( -5.41 = -4.85 + -0.56)
| Location | 15,588,802 – 15,588,859 |
|---|---|
| Length | 57 |
| Sequences | 5 |
| Columns | 61 |
| Reading direction | reverse |
| Mean pairwise identity | 68.07 |
| Shannon entropy | 0.56540 |
| G+C content | 0.27158 |
| Mean single sequence MFE | -7.36 |
| Consensus MFE | -4.65 |
| Energy contribution | -4.09 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.890767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
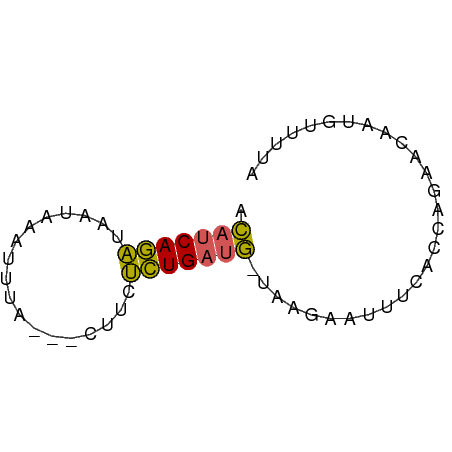
>dm3.chr3L 15588802 57 - 24543557 ACAUCAGAUAAUAAAUUUA---CUUCUCUGAUG-UAAGAAUUUCACCAGAACAAUGUUUUA ((((((((...........---....)))))))-).......................... ( -7.96, z-score = -1.04, R) >droSim1.chr3L 14928751 57 - 22553184 ACAUCAGAUAAUAAAUUUA---CUGCUCUGAUG-UAAGAAUUUCACCAGAACAAUUAUUUA ((((((((...........---....)))))))-).......................... ( -7.96, z-score = -1.67, R) >droYak2.chr3L 18136565 60 - 24197627 ACAUCAGACAAUAAAUUCAAUUCUUCUCUGAUG-UAAAAAUUUCACCAGAACAACGUUUUA ((((((((.((.((......)).)).)))))))-).......................... ( -7.90, z-score = -1.82, R) >droEre2.scaffold_4784 17847193 57 - 25762168 ACAUCAGAUAAUACAUUUACUUCUACUCUGAUG-UAAGAAUUUUACCAGAAGGUUGUA--- ((((((((..................)))))))-).........(((....)))....--- ( -8.97, z-score = -0.65, R) >droVir3.scaffold_13049 2766615 52 + 25233164 -UUACAAUUUUUAGGUUAA----UUCAUUGACACCAAGAAGUCCA----AAUGAUAUAUUA -..................----.((((((((........)))..----)))))....... ( -4.00, z-score = 0.23, R) >consensus ACAUCAGAUAAUAAAUUUA___CUUCUCUGAUG_UAAGAAUUUCACCAGAACAAUGUUUUA .(((((((..................)))))))............................ ( -4.65 = -4.09 + -0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:29:15 2011