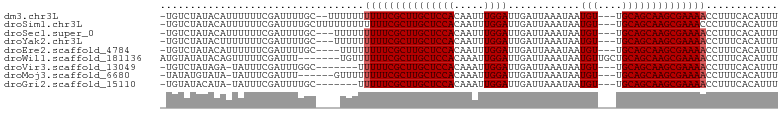
| Sequence ID | dm3.chr3L |
|---|---|
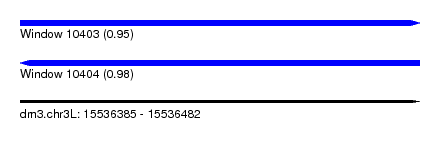
| Location | 15,536,385 – 15,536,482 |
| Length | 97 |
| Max. P | 0.979936 |

| Location | 15,536,385 – 15,536,482 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 90.84 |
| Shannon entropy | 0.18060 |
| G+C content | 0.31544 |
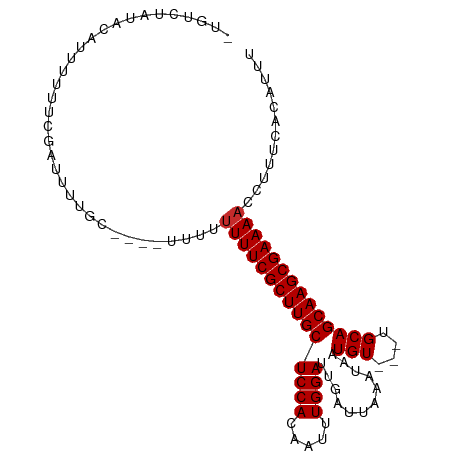
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -17.49 |
| Energy contribution | -17.60 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
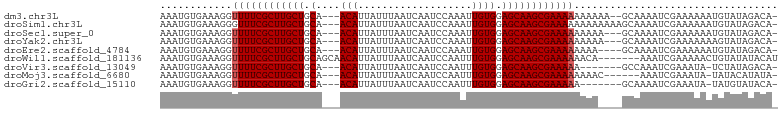
>dm3.chr3L 15536385 97 + 24543557 AAAUGUGAAAGGUUUUCGCUUGCUGCA---ACAUUAUUUAAUCAAUCCAAAUUGUGGAGCAAGCGAAAAAAAAAA--GCAAAAUCGAAAAAAUGUAUAGACA- ...(((......((((((((((((.((---.((...................)))).))))))))))))......--)))......................- ( -22.31, z-score = -2.77, R) >droSim1.chr3L 14877600 99 + 22553184 AAAUGUGAAAGGGUUUCGCUUGCUGCA---ACAUUAUUUAAUCAAUCCAAAUUGUGGAGCAAGCGAAAAAAAAAAAAGCAAAAUCGAAAAAAUGUAUAGACA- ...(((.......(((((((((((.((---.((...................)))).))))))))))).........)))......................- ( -20.10, z-score = -1.92, R) >droSec1.super_0 7645874 96 + 21120651 AAAUGUGAAAGGUUUUCGCUUGCUGCA---ACAUUAUUUAAUCAAUCCAAAUUGUGGAGCAAGCGAAAAAAAAA---GCAAAAUCGAAAAAAUGUAUAGACA- ...(((......((((((((((((.((---.((...................)))).)))))))))))).....---)))......................- ( -21.91, z-score = -2.59, R) >droYak2.chr3L 18052281 96 + 24197627 AAAUGUGAAAGGUUUUCGCUUGCUGCA---ACAUUAUUUAAUCAAUCCAAAUUGUGGAGCAAGCGAAAAAAAAA---GCAAAAUCGAAAAAAAGUAUAGACA- ...(((......((((((((((((.((---.((...................)))).)))))))))))).....---)))......................- ( -21.91, z-score = -2.75, R) >droEre2.scaffold_4784 17794083 95 + 25762168 AAAUGUGAAAGGUUUUCGCUUGCUGCA---ACAUUAUUUAAUCAAUCCAAAUUGUGGAGCAAGCGAAAAAAAA----GCAAAAUCGAAAAAAUGUAUAGACA- ...(((......((((((((((((.((---.((...................)))).))))))))))))....----)))......................- ( -21.51, z-score = -2.42, R) >droWil1.scaffold_181136 1873707 96 + 2313701 AAAUGUGAAAGGUUUUCGCUUGCUGCAGCAACAUUAUUUAAUCAAUCCAAUUUGUGGAGCAAGCGAAAAAACA-------AAAUCGAAAAACUGUAUAUACAU ..(((((.....((((((((((((.((.(((....................))))).)))))))))))).(((-------............)))...))))) ( -20.25, z-score = -1.78, R) >droVir3.scaffold_13049 20313579 91 + 25233164 AAAUGUGAAAGGUUUUCGCUUGCUGCA---ACAUUAUUUAAUCAAUCCAAUUUGUGGAGCAAGCGAAAAA-------GCCAAAUCGAAAUA-UCUAUAGACA- ..........((((((((((((((.((---.((...................)))).)))))))))))..-------)))...........-..........- ( -22.01, z-score = -2.44, R) >droMoj3.scaffold_6680 8026775 92 + 24764193 AAAUGUGAAAGGUUUUCGCUUGCUGCA---ACAUUAUUUAAUCAAUCCAAUUUGUGGAGCAAGCGAAAAAAAAC------AAAUCGAAAUA-UAUACAUAUA- ...(((......((((((((((((.((---.((...................)))).))))))))))))...))------)..........-..........- ( -20.81, z-score = -2.70, R) >droGri2.scaffold_15110 7978280 91 - 24565398 AAAUGUGAAAGGUUUUCGCUUGCUGCA---ACAUUAUUUAAUCAAUCCAAUUUGUGGAGCAAGCGAAAAA-------GCAAAAUCGAAAUA-UAUGUAUACA- ...(((......((((((((((((.((---.((...................)))).)))))))))))).-------)))...........-..........- ( -20.31, z-score = -1.84, R) >consensus AAAUGUGAAAGGUUUUCGCUUGCUGCA___ACAUUAUUUAAUCAAUCCAAAUUGUGGAGCAAGCGAAAAAAAA____GCAAAAUCGAAAAAAUGUAUAGACA_ ............((((((((((((.((........(((.....)))........)).)))))))))))).................................. (-17.49 = -17.60 + 0.11)
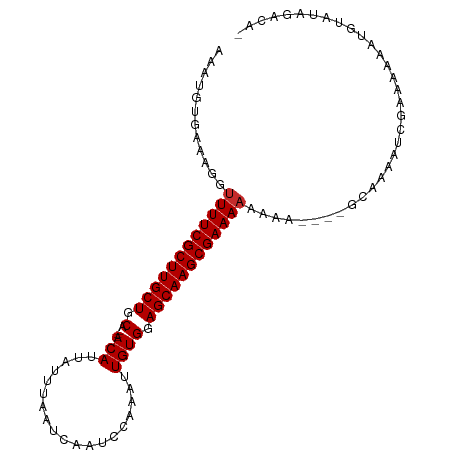
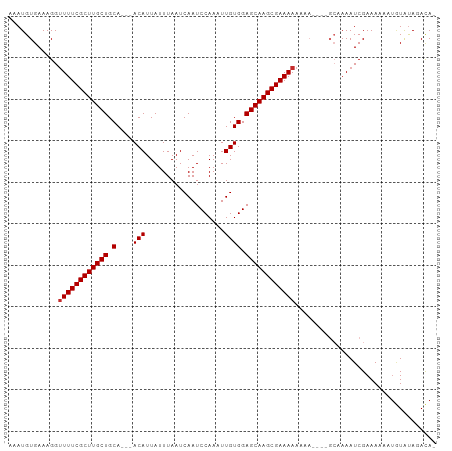
| Location | 15,536,385 – 15,536,482 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 90.84 |
| Shannon entropy | 0.18060 |
| G+C content | 0.31544 |
| Mean single sequence MFE | -18.49 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.51 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15536385 97 - 24543557 -UGUCUAUACAUUUUUUCGAUUUUGC--UUUUUUUUUUCGCUUGCUCCACAAUUUGGAUUGAUUAAAUAAUGU---UGCAGCAAGCGAAAACCUUUCACAUUU -.........................--......(((((((((((((.(((((((((.....))))))..)))---.).))))))))))))............ ( -18.30, z-score = -1.91, R) >droSim1.chr3L 14877600 99 - 22553184 -UGUCUAUACAUUUUUUCGAUUUUGCUUUUUUUUUUUUCGCUUGCUCCACAAUUUGGAUUGAUUAAAUAAUGU---UGCAGCAAGCGAAACCCUUUCACAUUU -..................................((((((((((((.(((((((((.....))))))..)))---.).)))))))))))............. ( -17.40, z-score = -1.78, R) >droSec1.super_0 7645874 96 - 21120651 -UGUCUAUACAUUUUUUCGAUUUUGC---UUUUUUUUUCGCUUGCUCCACAAUUUGGAUUGAUUAAAUAAUGU---UGCAGCAAGCGAAAACCUUUCACAUUU -.........................---.....(((((((((((((.(((((((((.....))))))..)))---.).))))))))))))............ ( -18.30, z-score = -1.88, R) >droYak2.chr3L 18052281 96 - 24197627 -UGUCUAUACUUUUUUUCGAUUUUGC---UUUUUUUUUCGCUUGCUCCACAAUUUGGAUUGAUUAAAUAAUGU---UGCAGCAAGCGAAAACCUUUCACAUUU -.........................---.....(((((((((((((.(((((((((.....))))))..)))---.).))))))))))))............ ( -18.30, z-score = -2.00, R) >droEre2.scaffold_4784 17794083 95 - 25762168 -UGUCUAUACAUUUUUUCGAUUUUGC----UUUUUUUUCGCUUGCUCCACAAUUUGGAUUGAUUAAAUAAUGU---UGCAGCAAGCGAAAACCUUUCACAUUU -.........................----....(((((((((((((.(((((((((.....))))))..)))---.).))))))))))))............ ( -18.30, z-score = -1.88, R) >droWil1.scaffold_181136 1873707 96 - 2313701 AUGUAUAUACAGUUUUUCGAUUU-------UGUUUUUUCGCUUGCUCCACAAAUUGGAUUGAUUAAAUAAUGUUGCUGCAGCAAGCGAAAACCUUUCACAUUU ((((....((((..........)-------))).((((((((((((.(((((.....(((.....)))....))).)).))))))))))))......)))).. ( -20.00, z-score = -1.58, R) >droVir3.scaffold_13049 20313579 91 - 25233164 -UGUCUAUAGA-UAUUUCGAUUUGGC-------UUUUUCGCUUGCUCCACAAAUUGGAUUGAUUAAAUAAUGU---UGCAGCAAGCGAAAACCUUUCACAUUU -((((....))-))....((...((.-------.((((((((((((.((...((((...........))))..---)).))))))))))))))..))...... ( -21.00, z-score = -2.11, R) >droMoj3.scaffold_6680 8026775 92 - 24764193 -UAUAUGUAUA-UAUUUCGAUUU------GUUUUUUUUCGCUUGCUCCACAAAUUGGAUUGAUUAAAUAAUGU---UGCAGCAAGCGAAAACCUUUCACAUUU -..........-...........------.....((((((((((((.((...((((...........))))..---)).))))))))))))............ ( -17.40, z-score = -1.43, R) >droGri2.scaffold_15110 7978280 91 + 24565398 -UGUAUACAUA-UAUUUCGAUUUUGC-------UUUUUCGCUUGCUCCACAAAUUGGAUUGAUUAAAUAAUGU---UGCAGCAAGCGAAAACCUUUCACAUUU -..........-..............-------.((((((((((((.((...((((...........))))..---)).))))))))))))............ ( -17.40, z-score = -1.49, R) >consensus _UGUCUAUACAUUUUUUCGAUUUUGC____UUUUUUUUCGCUUGCUCCACAAUUUGGAUUGAUUAAAUAAUGU___UGCAGCAAGCGAAAACCUUUCACAUUU ..................................(((((((((((((((.....))))............(((....))))))))))))))............ (-18.40 = -18.51 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:29:09 2011