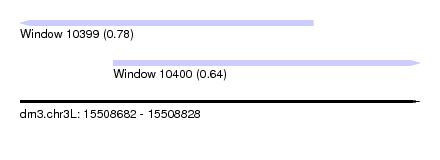
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,508,682 – 15,508,828 |
| Length | 146 |
| Max. P | 0.777913 |

| Location | 15,508,682 – 15,508,789 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.09 |
| Shannon entropy | 0.35725 |
| G+C content | 0.37711 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -15.16 |
| Energy contribution | -19.48 |
| Covariance contribution | 4.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
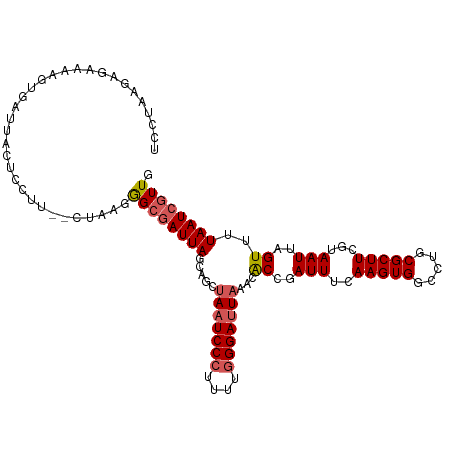
>dm3.chr3L 15508682 107 - 24543557 AUCGGUGUUUAAUCCCAAAAGGGAUUUACUGCUAAUCGCCCUUAGGAAAGAAGUAAUCACUUUUGUCUGAGGAAAAAUUAGGUGCAGUUUGU-AAGUAUCUGACUAAU ..(((((...((((((....)))))))))))........(((((((..((((((....)))))).))))))).....((((((((.......-..))))))))..... ( -27.40, z-score = -2.01, R) >droSim1.chr3L 14851525 105 - 22553184 AUCGGUGUUUAAUCCCAAAAGGGAUUAGCUGCUAAUCGCCCUUAG--AAGGAGUAAUCACUUUUCGCUUAGGAAAAAUUAGGUGCAGUUGGU-AAGUACCUGAUAAAU ...(((..((((((((....))))))))..)))......(((.((--.((((((....))))))..)).)))....(((((((((.......-..))))))))).... ( -28.20, z-score = -1.77, R) >droSec1.super_0 7618154 106 - 21120651 AUCGGUGUUUAAUCCCAAAAGGGAUUAGCUGCUAAUCGCCCAUAG--AAGGAGUAAUCACUUUUCGCUUAGGAAAAAUUAAGUGCAGUUGGUAAAGUAUCUGAUAAAU (((((...((((((((....)))))))).(((((((.((....((--(((((....)).))))).((((((......)))))))).)))))))......))))).... ( -27.10, z-score = -2.18, R) >droYak2.chr3L 18024156 83 - 24197627 AUCGGUGUUUAUUCCCAAAAGGGAUUA---GCUAAUGGCCCUUAG--AAGGAGUAAACACUUUUCUACGAGUCAAAAGUACAUGAAGU-------------------- ...((((((((((((...(((((.(((---.....))))))))..--..))))))))))))...........................-------------------- ( -21.20, z-score = -1.97, R) >consensus AUCGGUGUUUAAUCCCAAAAGGGAUUAGCUGCUAAUCGCCCUUAG__AAGGAGUAAUCACUUUUCGCUGAGGAAAAAUUAGGUGCAGUUGGU_AAGUAUCUGAUAAAU ...((((.((((((((....)))))))).))))......((((((...((((((....))))))..)))))).....((((((((..........))))))))..... (-15.16 = -19.48 + 4.31)



| Location | 15,508,716 – 15,508,828 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.98 |
| Shannon entropy | 0.37031 |
| G+C content | 0.40654 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -16.72 |
| Energy contribution | -17.60 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.635498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15508716 112 + 24543557 UCCUCAGACAAAAGUGAUUACUUCUUUCCUAAGGGCGAUUAGCAGUAAAUCCCUUUUGGGAUUAAACACCGAUUUCAAGUGGCCUGCGCUUCGUAAUUAGUUUUAAUCGUUG ......(((..((((....)))).....((((..((((...((((..((((((....))))))...(((.........)))..))))...))))..))))........))). ( -24.70, z-score = -0.82, R) >droSim1.chr3L 14851559 110 + 22553184 UCCUAAGCGAAAAGUGAUUACUCCUU--CUAAGGGCGAUUAGCAGCUAAUCCCUUUUGGGAUUAAACACCGAUUUCAAGUGGCCUGCGCUUCGUAAUUAGUUUUAAUCAUUG ............((((((((......--((((..((((...((((((((((((....)))))))..(((.........)))).))))...))))..))))...)))))))). ( -30.20, z-score = -2.34, R) >droSec1.super_0 7618189 110 + 21120651 UCCUAAGCGAAAAGUGAUUACUCCUU--CUAUGGGCGAUUAGCAGCUAAUCCCUUUUGGGAUUAAACACCGAUUUCAAGUGGCCUGCGCUUCGUAAUUAGUUUUAAUCGUUG .....((((((((.((((((((((..--....))).((...((((((((((((....)))))))..(((.........)))).))))...))))))))).)))...))))). ( -29.70, z-score = -2.03, R) >droYak2.chr3L 18024171 107 + 24197627 GACUCGUAGAAAAGUGUUUACUCCUU--CUAAGGGCCAUUAGC---UAAUCCCUUUUGGGAAUAAACACCGAUUUCAAGUGGUCUGCGCUUCAUAAUUAGUUUUAAUCGUUG ((..((((((...(((((((.((((.--..(((((........---....)))))..)))).))))))).(....)......))))))..)).................... ( -25.10, z-score = -1.69, R) >droEre2.scaffold_4784 17766542 90 + 25762168 ---------------------GCCUCGGAGAAAAGCGAUUA-CUCCUAAUCCAUUUUAGGAUUAUACGCCGAUUUCAAGUGGCCUGCGCUUCAUAAUUAGUUUUAAUCGUUG ---------------------............((((((((-.((((((......))))))......((..(((..(((((.....)))))...)))..))..)))))))). ( -18.20, z-score = -0.56, R) >consensus UCCUAAGAGAAAAGUGAUUACUCCUU__CUAAGGGCGAUUAGCAGCUAAUCCCUUUUGGGAUUAAACACCGAUUUCAAGUGGCCUGCGCUUCGUAAUUAGUUUUAAUCGUUG .................................(((((((((....(((((((....)))))))...((..(((..(((((.....)))))...)))..)).))))))))). (-16.72 = -17.60 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:29:06 2011