
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,484,625 – 15,484,724 |
| Length | 99 |
| Max. P | 0.750964 |

| Location | 15,484,625 – 15,484,724 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 84.40 |
| Shannon entropy | 0.31248 |
| G+C content | 0.32337 |
| Mean single sequence MFE | -14.81 |
| Consensus MFE | -13.07 |
| Energy contribution | -12.85 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.750964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
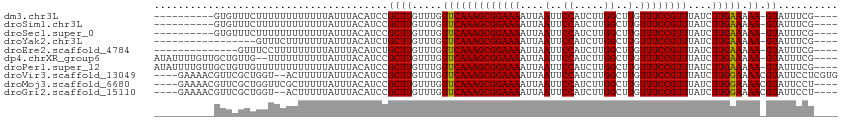
>dm3.chr3L 15484625 99 + 24543557 ----CGAAAUAC-UUUUUCAAGAUAAACGGAAACAAGCCAAGAUGGAAUUAAUUUUCCGCUUUGAACAAACAAGCGGAUGUAAAUAAAAAAAAAAAAGAAACAC---------- ----.......(-(((((..........(....)...((.....)).........(((((((.(......))))))))..............))))))......---------- ( -13.60, z-score = -1.46, R) >droSim1.chr3L 14824567 99 + 22553184 ----CGAAAUAC-UUUUUCAAGAUAAACGGAAACAAGCCAAGAUGGAAUUAAUUUUCCGCUUUGAACAAACAAGCGGAUGUAAAUAAAAAAAAAAAAGAAACAC---------- ----.......(-(((((..........(....)...((.....)).........(((((((.(......))))))))..............))))))......---------- ( -13.60, z-score = -1.46, R) >droSec1.super_0 7593187 99 + 21120651 ----CGAAAUAC-UUUUUCAAGAUAAACGGAAACAAGCCAAGAUGGAAUUAAUUUUCCGCUUUGAACAAACAAGCGGAUGUAAAUAAAAAAAAAAAAGAAACAC---------- ----.......(-(((((..........(....)...((.....)).........(((((((.(......))))))))..............))))))......---------- ( -13.60, z-score = -1.46, R) >droYak2.chr3L 17999166 92 + 24197627 ----CGAAAUAC-UUUUUCAAGAUAAACGGAAACAAGCCAAGAUGGAAUUAAUUUUCCGCUUUGAACAAACAAGCAGAUGUAAAUAAAAAAAGAAAC----------------- ----.......(-(((((..........(....)....((((.(((((......))))).)))).......................))))))....----------------- ( -11.40, z-score = -0.96, R) >droEre2.scaffold_4784 17739714 95 + 25762168 ----CGAAAUAC-UUUUUCAAGAUAAACGGAAACAAGCCAAGAUGGAAUUAAUUUUCCGCUUUGAACAAACAAGCAGAUGUAAAUAAAAAAAAAGGAAAC-------------- ----.((((...-..)))).........(....)...((.....((((......))))((((.(......)))))...................))....-------------- ( -11.80, z-score = -0.82, R) >dp4.chrXR_group6 13043935 107 - 13314419 ----CGAAAUAC-UUUUUCAAGAUAAACGGAAACAAGCCAAGAUGGAAUUAAUUUUCCGCUUUGAACAAACAAGCGGAUGUAAAUAAAAAAAAAA--CAACAGCAACAAAAUAU ----.((((...-..)))).........(....)...((.....)).........(((((((.(......)))))))).................--................. ( -13.10, z-score = -1.16, R) >droPer1.super_12 1457861 109 - 2414086 ----CGAAAUAC-UUUUUCAAGAUAAACGGAAACAAGCCAAGAUGGAAUUAAUUUUCCGCUUUGAACAAACAAGCGGAUGUAAAUAAAAAAAAAAAACAACAGCAACAAAAUAU ----.((((...-..)))).........(....)...((.....)).........(((((((.(......)))))))).................................... ( -13.10, z-score = -1.15, R) >droVir3.scaffold_13049 20261953 108 + 25233164 CACGAGGAAUACGUUUUCCAAGAUAAACGGAAACAAGCCAAGAUGGAAUUAAUUUUCCGCUUUGAACAAACAAGCGGAUGUAAAUAAAAAGU--ACCAGCGAACGUUUUC---- ......(((.((((((((((........(....).........))))........(((((((.(......))))))))..............--......)))))).)))---- ( -18.73, z-score = -0.56, R) >droMoj3.scaffold_6680 7965893 106 + 24764193 ----AGGAAUACGUUUUCCAAGAUAAACGGAAACAAGCCAAGAUGGAAUUAAUUUUCCGCUUUGAACAAACAAGCGGAUGUAAAUAAAAAGCGAACCAGCGAACGUUUUC---- ----..(((.((((((((((........(....).........))))........(((((((.(......))))))))............((......)))))))).)))---- ( -20.43, z-score = -1.02, R) >droGri2.scaffold_15110 4658737 104 - 24565398 ----AGGAAUACGUUUUCCAAGAUAAACGGAAACAAGCCAAGAUGGAAUUAAUUUUCCGCUUUGAACAAACAAGCGGAUGUAAAUAAAAAGU--ACCAGCGAACGUUUUC---- ----..(((.((((((((((........(....).........))))........(((((((.(......))))))))..............--......)))))).)))---- ( -18.73, z-score = -0.78, R) >consensus ____CGAAAUAC_UUUUUCAAGAUAAACGGAAACAAGCCAAGAUGGAAUUAAUUUUCCGCUUUGAACAAACAAGCGGAUGUAAAUAAAAAAAAAAAAAAAAAAC__________ .....((((......)))).........(....)...((.....)).........(((((((.(......)))))))).................................... (-13.07 = -12.85 + -0.22)


| Location | 15,484,625 – 15,484,724 |
|---|---|
| Length | 99 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.40 |
| Shannon entropy | 0.31248 |
| G+C content | 0.32337 |
| Mean single sequence MFE | -16.62 |
| Consensus MFE | -12.49 |
| Energy contribution | -12.07 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.550815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15484625 99 - 24543557 ----------GUGUUUCUUUUUUUUUUUUAUUUACAUCCGCUUGUUUGUUCAAAGCGGAAAAUUAAUUCCAUCUUGGCUUGUUUCCGUUUAUCUUGAAAAA-GUAUUUCG---- ----------.............................((((.....(((((((((((((....(..((.....))..).))))))))....))))).))-))......---- ( -13.70, z-score = -1.08, R) >droSim1.chr3L 14824567 99 - 22553184 ----------GUGUUUCUUUUUUUUUUUUAUUUACAUCCGCUUGUUUGUUCAAAGCGGAAAAUUAAUUCCAUCUUGGCUUGUUUCCGUUUAUCUUGAAAAA-GUAUUUCG---- ----------.............................((((.....(((((((((((((....(..((.....))..).))))))))....))))).))-))......---- ( -13.70, z-score = -1.08, R) >droSec1.super_0 7593187 99 - 21120651 ----------GUGUUUCUUUUUUUUUUUUAUUUACAUCCGCUUGUUUGUUCAAAGCGGAAAAUUAAUUCCAUCUUGGCUUGUUUCCGUUUAUCUUGAAAAA-GUAUUUCG---- ----------.............................((((.....(((((((((((((....(..((.....))..).))))))))....))))).))-))......---- ( -13.70, z-score = -1.08, R) >droYak2.chr3L 17999166 92 - 24197627 -----------------GUUUCUUUUUUUAUUUACAUCUGCUUGUUUGUUCAAAGCGGAAAAUUAAUUCCAUCUUGGCUUGUUUCCGUUUAUCUUGAAAAA-GUAUUUCG---- -----------------.....................(((((.....(((((((((((((....(..((.....))..).))))))))....))))).))-))).....---- ( -14.40, z-score = -1.70, R) >droEre2.scaffold_4784 17739714 95 - 25762168 --------------GUUUCCUUUUUUUUUAUUUACAUCUGCUUGUUUGUUCAAAGCGGAAAAUUAAUUCCAUCUUGGCUUGUUUCCGUUUAUCUUGAAAAA-GUAUUUCG---- --------------........................(((((.....(((((((((((((....(..((.....))..).))))))))....))))).))-))).....---- ( -14.40, z-score = -1.66, R) >dp4.chrXR_group6 13043935 107 + 13314419 AUAUUUUGUUGCUGUUG--UUUUUUUUUUAUUUACAUCCGCUUGUUUGUUCAAAGCGGAAAAUUAAUUCCAUCUUGGCUUGUUUCCGUUUAUCUUGAAAAA-GUAUUUCG---- (((((((...((.(.((--(.............))).).)).......(((((((((((((....(..((.....))..).))))))))....))))))))-))))....---- ( -14.62, z-score = -0.62, R) >droPer1.super_12 1457861 109 + 2414086 AUAUUUUGUUGCUGUUGUUUUUUUUUUUUAUUUACAUCCGCUUGUUUGUUCAAAGCGGAAAAUUAAUUCCAUCUUGGCUUGUUUCCGUUUAUCUUGAAAAA-GUAUUUCG---- (((((((...((.(.(((...............))).).)).......(((((((((((((....(..((.....))..).))))))))....))))))))-))))....---- ( -14.46, z-score = -0.56, R) >droVir3.scaffold_13049 20261953 108 - 25233164 ----GAAAACGUUCGCUGGU--ACUUUUUAUUUACAUCCGCUUGUUUGUUCAAAGCGGAAAAUUAAUUCCAUCUUGGCUUGUUUCCGUUUAUCUUGGAAAACGUAUUCCUCGUG ----(((.(((((.((..(.--..............(((((((.........)))))))..............)..))....(((((.......)))))))))).)))...... ( -21.45, z-score = -1.36, R) >droMoj3.scaffold_6680 7965893 106 - 24764193 ----GAAAACGUUCGCUGGUUCGCUUUUUAUUUACAUCCGCUUGUUUGUUCAAAGCGGAAAAUUAAUUCCAUCUUGGCUUGUUUCCGUUUAUCUUGGAAAACGUAUUCCU---- ----(((.(((((.((..(...(((((..((..(((......)))..))..)))))((((......))))...)..))....(((((.......)))))))))).)))..---- ( -24.30, z-score = -2.25, R) >droGri2.scaffold_15110 4658737 104 + 24565398 ----GAAAACGUUCGCUGGU--ACUUUUUAUUUACAUCCGCUUGUUUGUUCAAAGCGGAAAAUUAAUUCCAUCUUGGCUUGUUUCCGUUUAUCUUGGAAAACGUAUUCCU---- ----(((.(((((.((..(.--..............(((((((.........)))))))..............)..))....(((((.......)))))))))).)))..---- ( -21.45, z-score = -1.68, R) >consensus __________GUUCUUUUUUUUUUUUUUUAUUUACAUCCGCUUGUUUGUUCAAAGCGGAAAAUUAAUUCCAUCUUGGCUUGUUUCCGUUUAUCUUGAAAAA_GUAUUUCG____ .............................((..(((......)))..))...(((((((((....(..((.....))..).))))))))).....((((......))))..... (-12.49 = -12.07 + -0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:28:59 2011