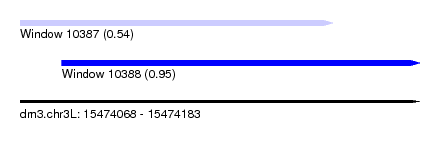
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,474,068 – 15,474,183 |
| Length | 115 |
| Max. P | 0.947073 |

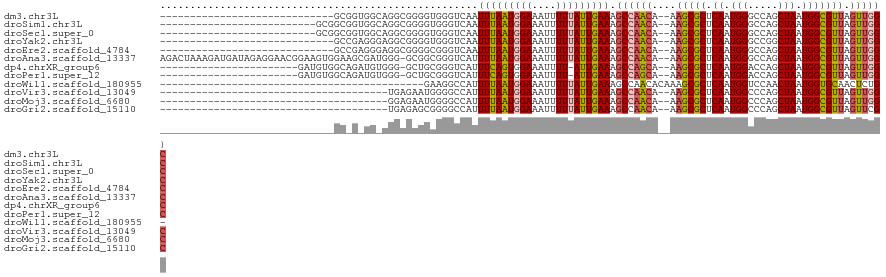
| Location | 15,474,068 – 15,474,158 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 77.36 |
| Shannon entropy | 0.41376 |
| G+C content | 0.47905 |
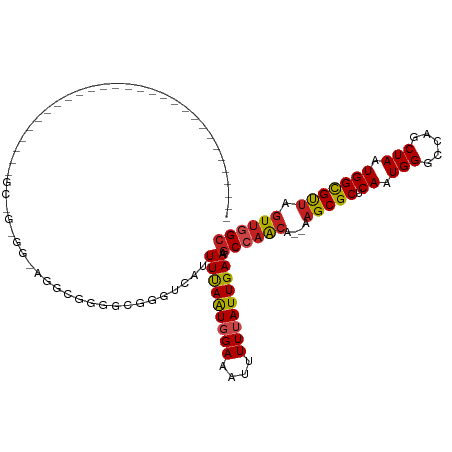
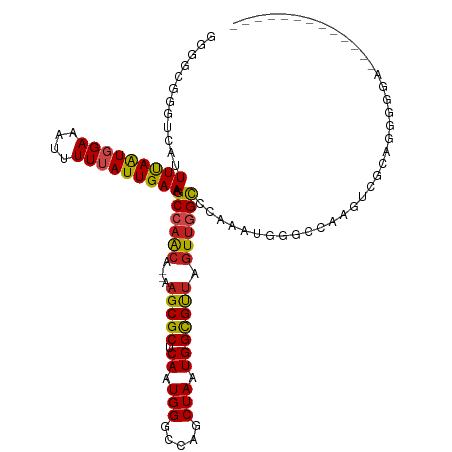
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -17.61 |
| Energy contribution | -17.71 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
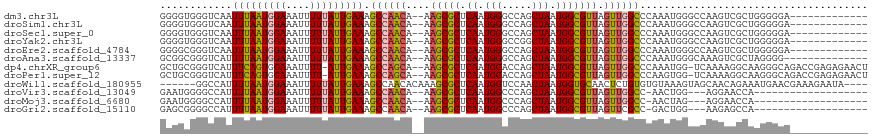
>dm3.chr3L 15474068 90 + 24543557 -----------------------------GCGGUGGCAGGCGGGGUGGGUCAAUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGGCCAGCUAAUGGCGUUAGUUGGC -----------------------------((.(((((.(((.......)))..(((((((((....))))))))).))).)).--..)).........((((((((((...)))))))))) ( -25.40, z-score = -0.60, R) >droSim1.chr3L 14813837 93 + 22553184 --------------------------GCGGCGGUGGCAGGCGGGGUGGGUCAAUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGGCCAGCUAAUGGCGUUAGUUGGC --------------------------(((.(.(((((.(((.......)))..(((((((((....))))))))).))).)).--..)))).......((((((((((...)))))))))) ( -26.90, z-score = -0.39, R) >droSec1.super_0 7582748 93 + 21120651 --------------------------GCGGCGGUGGCAGGCGGGGUGGGUCAAUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGGCCAGCUAAUGGCGUUAGUUGGC --------------------------(((.(.(((((.(((.......)))..(((((((((....))))))))).))).)).--..)))).......((((((((((...)))))))))) ( -26.90, z-score = -0.39, R) >droYak2.chr3L 17988842 90 + 24197627 -----------------------------GCCGAGGGAGGCGGGGUGGGUCAAUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGGCCGGCUAAUGGCGUUAGUUGGC -----------------------------.(((...(((((..(((...(((((..(((....)))...)))))..)))....--..)).)))..)))((((((((((...)))))))))) ( -28.00, z-score = -1.36, R) >droEre2.scaffold_4784 17728601 90 + 25762168 -----------------------------GCCGAGGGAGGCGGGGCGGGUCAAUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGGCCAGCUAAUGGCGUUAGUUGGC -----------------------------.(((...(((((..(((...(((((..(((....)))...)))))..)))....--..)).)))..)))((((((((((...)))))))))) ( -30.40, z-score = -2.06, R) >droAna3.scaffold_13337 5906470 118 - 23293914 AGACUAAAGAUGAUAGAGGAACGGAAGUGGAAGCGAUGGG-GCGGCGGGUCAUUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGGCCAGCUAAUGGCGUUAGUUGGC ...(((..............((....))........((((-((.(..(((..((((((((((....)))))))))))))..).--..)).)))).)))((((((((((...)))))))))) ( -29.60, z-score = -1.01, R) >dp4.chrXR_group6 13028708 94 - 13314419 -----------------------GAUGUGGCAGAUGUGGG-GCUGCGGGUCAUUUCAGUGGAAAUUUU-AUUGAAAGCCAGCA--AAGCGCUCAAUGGACCAGCUAAUGGCGUUAGUUGGC -----------------------(((.((.(((.......-.))))).))).((((((((((...)))-)))))))((((((.--.(((((.((.(((.....))).))))))).)))))) ( -28.80, z-score = -0.80, R) >droPer1.super_12 1446516 94 - 2414086 -----------------------GAUGUGGCAGAUGUGGG-GCUGCGGGUCAUUUCAGUGGAAAUUUU-AUUGAAAGCCAGCA--AAGCGCUCAAUGGACCAGCUAAUGGCGUUAGUUGGC -----------------------(((.((.(((.......-.))))).))).((((((((((...)))-)))))))((((((.--.(((((.((.(((.....))).))))))).)))))) ( -28.80, z-score = -0.80, R) >droWil1.scaffold_180955 2396055 76 - 2875958 --------------------------------------------GAAGGCCAUUUUAAUGGAAAUUUUUAUUGAAAGCCAACACAAAGCGCUCAAUGGUCCAACUAAUGGUGCAACUCUG- --------------------------------------------...(((..((((((((((....)))))))))))))........((((.((.(((.....))).)))))).......- ( -17.40, z-score = -1.49, R) >droVir3.scaffold_13049 20250238 81 + 25233164 --------------------------------------UGAGAAUGGGGCCAUUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGCCCAGCUAAUGGCGUUAGUUGGC --------------------------------------(((((((....((((....))))..)))))))......((((((.--.(((((.((.((((...)))).))))))).)))))) ( -24.50, z-score = -1.67, R) >droMoj3.scaffold_6680 7954143 81 + 24764193 --------------------------------------GGAGAAUGGGGCCAUUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGCCCAGCUAAUGGCGUUAGUUGGC --------------------------------------.((((((....((((....))))..)))))).......((((((.--.(((((.((.((((...)))).))))))).)))))) ( -24.00, z-score = -1.38, R) >droGri2.scaffold_15110 4647645 81 - 24565398 --------------------------------------UGAGAGCGGGGCCAUUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGCCCAGCUAAUGGCGUUAGUUCGC --------------------------------------...((((.((((((((((((((((....)))))))).((((....--..).))).)))))))).((.....))....)))).. ( -23.90, z-score = -1.24, R) >consensus _____________________________GC_G_GG_AGGCGGGGCGGGUCAUUUUAAUGGAAAUUUUUAUUGAAAGCCAACA__AAGCGCUCAAUGGGCCAGCUAAUGGCGUUAGUUGGC .....................................................(((((((((....))))))))).((((((....(((((.((.(((.....))).))))))).)))))) (-17.61 = -17.71 + 0.10)
| Location | 15,474,080 – 15,474,183 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.82 |
| Shannon entropy | 0.45742 |
| G+C content | 0.47694 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -18.79 |
| Energy contribution | -18.73 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15474080 103 + 24543557 GGGGUGGGUCAAUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGGCCAGCUAAUGGCGUUAGUUGGCCCAAAUGGGCCAAGUCGCUGGGGGA------------- ..(((...(((((..(((....)))...)))))..)))..(.--.((((((((.(((((((((((((...)))))))))))))..)))))......)))..)...------------- ( -37.70, z-score = -2.17, R) >droSim1.chr3L 14813852 103 + 22553184 GGGGUGGGUCAAUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGGCCAGCUAAUGGCGUUAGUUGGCCCAAAUGGGCCAAGUCGCUGGGGGA------------- ..(((...(((((..(((....)))...)))))..)))..(.--.((((((((.(((((((((((((...)))))))))))))..)))))......)))..)...------------- ( -37.70, z-score = -2.17, R) >droSec1.super_0 7582763 103 + 21120651 GGGGUGGGUCAAUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGGCCAGCUAAUGGCGUUAGUUGGCCCAAAUGGGCCAAGUCGCUGGGGGA------------- ..(((...(((((..(((....)))...)))))..)))..(.--.((((((((.(((((((((((((...)))))))))))))..)))))......)))..)...------------- ( -37.70, z-score = -2.17, R) >droYak2.chr3L 17988854 103 + 24197627 GGGGUGGGUCAAUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGGCCGGCUAAUGGCGUUAGUUGGCCCAAAUGGGCCAAGUCGCUGGGGGA------------- ..(((...(((((..(((....)))...)))))..)))..(.--.((((((((.(((((((((((((...)))))))))))))..)))))......)))..)...------------- ( -36.90, z-score = -1.80, R) >droEre2.scaffold_4784 17728613 103 + 25762168 GGGGCGGGUCAAUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGGCCAGCUAAUGGCGUUAGUUGGCCCAAAUGGGCCAAGUCGCUGGGGGA------------- ..(((...(((((..(((....)))...)))))..)))..(.--.((((((((.(((((((((((((...)))))))))))))..)))))......)))..)...------------- ( -39.30, z-score = -2.44, R) >droAna3.scaffold_13337 5906510 102 - 23293914 GCGGCGGGUCAUUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGGCCAGCUAAUGGCGUUAGUUGGCCCAAAUGGGCAAAGUCGCUAGGGG-------------- (((((.(((..((((((((((....)))))))))))))....--....(((((.(((((((((((((...)))))))))))))..)))))...)))))......-------------- ( -42.90, z-score = -4.38, R) >dp4.chrXR_group6 13028725 114 - 13314419 GCUGCGGGUCAUUUCAGUGGAAAUUUU-AUUGAAAGCCAGCA--AAGCGCUCAAUGGACCAGCUAAUGGCGUUAGUUGGCCCAAAUGG-UCAAAAGGCAAGGGCAGACCGAGAGAACU (((((.(((..((((((((((...)))-)))))))))).)).--.))).(((..(((.(((((((((...))))))))).)))..(((-((.....((....)).))))).))).... ( -36.70, z-score = -1.52, R) >droPer1.super_12 1446533 114 - 2414086 GCUGCGGGUCAUUUCAGUGGAAAUUUU-AUUGAAAGCCAGCA--AAGCGCUCAAUGGACCAGCUAAUGGCGUUAGUUGGCCCAAGUGG-UCAAAAGGCAAGGGCAGACCGAGAGAACU (((((.(((..((((((((((...)))-)))))))))).)).--.))).(((..(((.(((((((((...))))))))).)))..(((-((.....((....)).))))).))).... ( -36.60, z-score = -1.31, R) >droWil1.scaffold_180955 2396058 108 - 2875958 ------GGCCAUUUUAAUGGAAAUUUUUAUUGAAAGCCAACACAAAGCGCUCAAUGGUCCAACUAAUGGUGCAACUCUGUGUGUAAAGUAGCAACAGAAAUGAACGAAAGAAUA---- ------(((..((((((((((....)))))))))))))........((((.((.(((.....))).))))))...(((((.(((......))))))))......(....)....---- ( -24.40, z-score = -1.39, R) >droVir3.scaffold_13049 20250241 93 + 25233164 GAAUGGGGCCAUUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGCCCAGCUAAUGGCGUUAGUUGGCC-AACUGG---AGGAACCA------------------- ...(((..((.((((((((((....))))))))))((((((.--.(((((.((.((((...)))).))))))).)))))).-......---.))..)))------------------- ( -31.20, z-score = -2.09, R) >droMoj3.scaffold_6680 7954146 93 + 24764193 GAAUGGGGCCAUUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGCCCAGCUAAUGGCGUUAGUUGGCC-AACUAG---AGGAACCA------------------- ...(((..((.((((((((((....))))))))))((((((.--.(((((.((.((((...)))).))))))).)))))).-......---.))..)))------------------- ( -31.20, z-score = -2.52, R) >droGri2.scaffold_15110 4647648 93 - 24565398 GAGCGGGGCCAUUUUAAUGGAAAUUUUUAUUGAAAGCCAACA--AAGCGCUCAAUGGCCCAGCUAAUGGCGUUAGUUCGCC-GACUGG---AAGAGCCA------------------- .(((.((((((((((((((((....)))))))).((((....--..).))).)))))))).)))..((((.((((((....-))))))---....))))------------------- ( -30.10, z-score = -1.47, R) >consensus GGGGCGGGUCAUUUUAAUGGAAAUUUUUAUUGAAAGCCAACA__AAGCGCUCAAUGGGCCAGCUAAUGGCGUUAGUUGGCCCAAAUGGGCCAAGUCGCAGGGGGA_____________ ............(((((((((....))))))))).((((((....(((((.((.(((.....))).))))))).))))))...................................... (-18.79 = -18.73 + -0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:28:55 2011