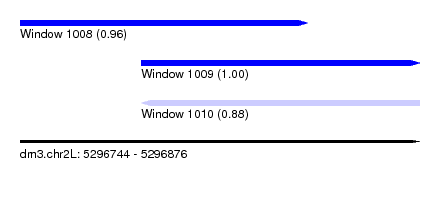
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,296,744 – 5,296,876 |
| Length | 132 |
| Max. P | 0.998817 |

| Location | 5,296,744 – 5,296,839 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 73.90 |
| Shannon entropy | 0.41925 |
| G+C content | 0.32935 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -13.73 |
| Energy contribution | -15.72 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
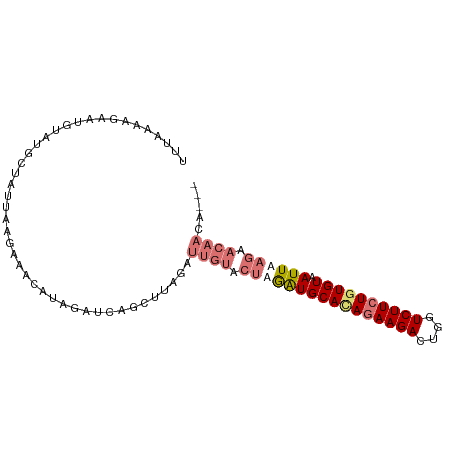
>dm3.chr2L 5296744 95 + 23011544 UUUCAAAGAAUGUGUGCUAUUACGAAACAUAGAUCAGCUUAGAUUGUACUAGAUGCAUAGAAGACUGGUCUUCUGUGUAAUUAAGAACAACACUG ...........((((.(((.(((((.....((.....))....))))).))).(((((((((((....)))))))))))..........)))).. ( -21.40, z-score = -1.20, R) >droSim1.chr2L_random 334773 92 + 909653 UUUAAAAGAAUGUAUGCUAUUAAGAAACAUAGAUCAGCUUAGAUUGUACUUGAUGCACAGAAGACUGGUCUUCUGUGUAAUUAACAACACUG--- ..........(((......(((((.......((((......))))...)))))(((((((((((....)))))))))))....)))......--- ( -19.60, z-score = -1.33, R) >droSec1.super_5 3375771 95 + 5866729 UUUAAAAGAAUGUAUGCUAUUAAGAAACAUAGAUCAGUUUAGAUUGUACUUGAUGCACAGAAGACUCGUCUUCUGUGUAAUUAAGAACAACCCUG ((((((...((.((((...........)))).))...))))))((((.((((((((((((((((....)))))))))).)))))).))))..... ( -26.00, z-score = -4.04, R) >droYak2.chr2L 8423655 85 - 22324452 --AAUCAUAUUAAGAACGAGUAAGAAACAUAUGC-----UAGAUUAUCCUAAGUGCAUAUAAGACUGCUCUUUUCUGUGAUUAUGAUCCACA--- --.((((((...((((.(((((......((((((-----(((......)))...)))))).....)))))..)))).....)))))).....--- ( -15.70, z-score = -1.23, R) >consensus UUUAAAAGAAUGUAUGCUAUUAAGAAACAUAGAUCAGCUUAGAUUGUACUAGAUGCACAGAAGACUGGUCUUCUGUGUAAUUAAGAACAACA___ ...........................................((((.((((((((((((((((....)))))))))).)))))).))))..... (-13.73 = -15.72 + 2.00)


| Location | 5,296,784 – 5,296,876 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 80.29 |
| Shannon entropy | 0.26818 |
| G+C content | 0.34543 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -19.37 |
| Energy contribution | -21.15 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.98 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.50 |
| SVM RNA-class probability | 0.998817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
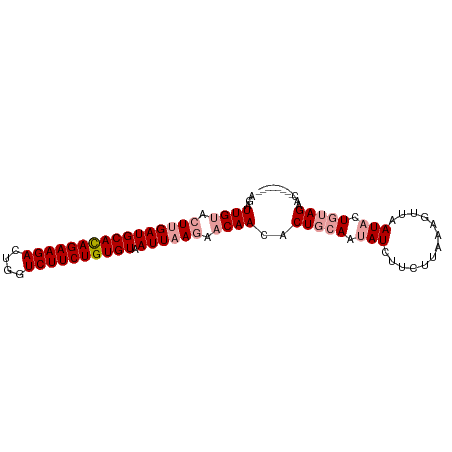
>dm3.chr2L 5296784 92 + 23011544 AGAUUGUACUAGAUGCAUAGAAGACUGGUCUUCUGUGUAAUUAAGAACAACACUGCAAUAUCAUCUUAAG--UCAUUCUUAAGAACUAUAGAAU ...((((.((.(((((((((((((....)))))))))).))).)).)))).............(((((((--.....))))))).......... ( -23.40, z-score = -2.48, R) >droSim1.chr2L_random 334813 83 + 909653 AGAUUGUACUUGAUGCACAGAAGACUGGUCUUCUGUGUAAUUAA---CAACACUGCAUUAUCUUGUUAAAGUUAAUACUGUAGAAC-------- ....(((..(((((((((((((((....)))))))))).)))))---..)))((((((((.(((....))).)))...)))))...-------- ( -22.70, z-score = -2.62, R) >droSec1.super_5 3375811 86 + 5866729 AGAUUGUACUUGAUGCACAGAAGACUCGUCUUCUGUGUAAUUAAGAACAACCCUGCAGUAUCUUCUUCAAGUUGAUACUGUAGAAC-------- ...((((.((((((((((((((((....)))))))))).)))))).))))..((((((((((..(.....)..))))))))))...-------- ( -36.40, z-score = -6.85, R) >consensus AGAUUGUACUUGAUGCACAGAAGACUGGUCUUCUGUGUAAUUAAGAACAACACUGCAAUAUCUUCUUAAAGUUAAUACUGUAGAAC________ ...((((.((((((((((((((((....)))))))))).)))))).))))..(((((.(((.............))).)))))........... (-19.37 = -21.15 + 1.78)

| Location | 5,296,784 – 5,296,876 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 80.29 |
| Shannon entropy | 0.26818 |
| G+C content | 0.34543 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -13.12 |
| Energy contribution | -15.57 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
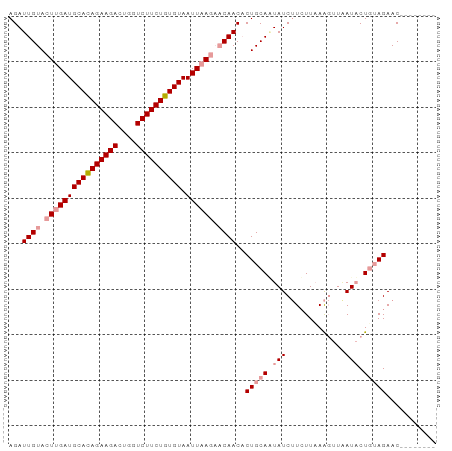
>dm3.chr2L 5296784 92 - 23011544 AUUCUAUAGUUCUUAAGAAUGA--CUUAAGAUGAUAUUGCAGUGUUGUUCUUAAUUACACAGAAGACCAGUCUUCUAUGCAUCUAGUACAAUCU ....(((...(((((((.....--)))))))..))).((((((((.((.....)).))))((((((....)))))).))))............. ( -18.90, z-score = -0.97, R) >droSim1.chr2L_random 334813 83 - 909653 --------GUUCUACAGUAUUAACUUUAACAAGAUAAUGCAGUGUUG---UUAAUUACACAGAAGACCAGUCUUCUGUGCAUCAAGUACAAUCU --------........(((((....(((((((.((......)).)))---))))...(((((((((....))))))))).....)))))..... ( -21.90, z-score = -3.19, R) >droSec1.super_5 3375811 86 - 5866729 --------GUUCUACAGUAUCAACUUGAAGAAGAUACUGCAGGGUUGUUCUUAAUUACACAGAAGACGAGUCUUCUGUGCAUCAAGUACAAUCU --------...((.(((((((..(.....)..))))))).))((((((.(((.....(((((((((....)))))))))....))).)))))). ( -29.00, z-score = -3.94, R) >consensus ________GUUCUACAGUAUCAACUUUAAGAAGAUAAUGCAGUGUUGUUCUUAAUUACACAGAAGACCAGUCUUCUGUGCAUCAAGUACAAUCU ...........((.(((((((...........))))))).)).(((((.(((.....(((((((((....)))))))))....))).))))).. (-13.12 = -15.57 + 2.45)

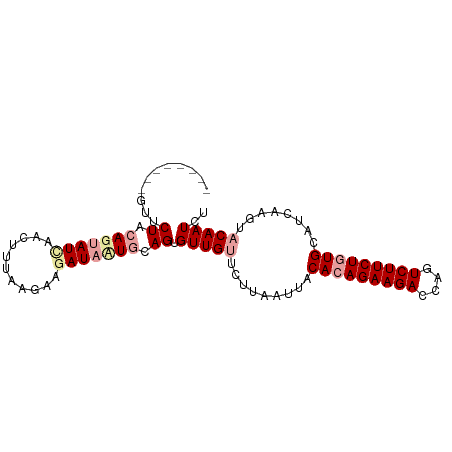
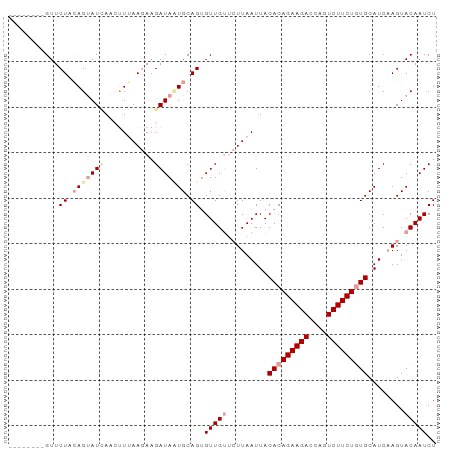
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:44 2011