
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,425,769 – 15,425,832 |
| Length | 63 |
| Max. P | 0.544454 |

| Location | 15,425,769 – 15,425,832 |
|---|---|
| Length | 63 |
| Sequences | 11 |
| Columns | 67 |
| Reading direction | forward |
| Mean pairwise identity | 65.90 |
| Shannon entropy | 0.68770 |
| G+C content | 0.36172 |
| Mean single sequence MFE | -13.56 |
| Consensus MFE | -2.43 |
| Energy contribution | -2.60 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.18 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.544454 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 15425769 63 + 24543557 UUUUUAUUUUUUUUCAUUCUCUUUUCC----CACUUUCGGUGUCUCGAUCAGAUACCAAAAGGAUGG ..........................(----((((((.(((((((.....))))))).))))..))) ( -12.10, z-score = -2.17, R) >droSec1.super_0 7532024 63 + 21120651 UUUUUAUUUUUUUUCAUUCUCUUUUCC----CACUUUCGGUGUCUCGAUCAGAUACCAAAAGGAUGG ..........................(----((((((.(((((((.....))))))).))))..))) ( -12.10, z-score = -2.17, R) >droEre2.scaffold_4784 17681873 59 + 25762168 ---UUUUUAUUUUUCAUUCUGUUUUCC----CACUUUCGGUGUCUCGAUCAGAUACC-AAAGGAUGG ---.......................(----((((((.(((((((.....)))))))-.)))).))) ( -13.50, z-score = -2.41, R) >droYak2.chr3L 17939933 59 + 24197627 ---UUUUUAUUUUUCAUUCUGUUUUCC----CACUUUCGGUGUCUCGAUCAGAUACC-AAAGGAUGG ---.......................(----((((((.(((((((.....)))))))-.)))).))) ( -13.50, z-score = -2.41, R) >droAna3.scaffold_13337 5863004 63 - 23293914 UUUUUUUUUUUGUCCGUUCUAUUUUUC----CACUUUUGGUGUCUCAAUCAGAUACCAAACGGAUGG .........(..((((((.........----......((((((((.....))))))))))))))..) ( -16.06, z-score = -3.96, R) >dp4.chrXR_group6 12981013 56 - 13314419 -------GUUUUUUUUUCGAUUUUUUC----GACUUUUGUUGUCUCAAUCAGAUAGCAGAAGGAUAG -------.........((((.....))----))((((((((((((.....))))))))))))..... ( -16.20, z-score = -4.20, R) >droPer1.super_12 1399987 56 - 2414086 -------GUUUUUUUUUCGAUUUUUUC----GACUUUUGUUGUCUCAAUCAGAUAGCAGAGGGAUAG -------.........((((.....))----))((((((((((((.....))))))))))))..... ( -15.90, z-score = -3.99, R) >droWil1.scaffold_180955 2339882 54 - 2875958 ---------UUUUUUUUUUUAUUUUUU----UUGGUUUAGCGUCUCAAUCAGAUAGCAAUGGGGCAA ---------..................----..........((((((............)))))).. ( -5.30, z-score = -0.10, R) >droVir3.scaffold_13049 20201730 66 + 25233164 UUCCAAUUUUUUGCUGCUCUGUUUUGUAGGACAGUUUUGGCGUCUCAAUCAGAUAGCGCAAACAGA- .........(((((.((((((..(((..((((.((....))))))))).))))..)))))))....- ( -13.30, z-score = -0.09, R) >droMoj3.scaffold_6680 7903852 67 + 24764193 UUUCUUUUUUUUCUAGCUGUGUUUUGUAGGACAGCUUUGGCGUCUCAAUCAGAUAGCGCAAACAGAA ..........((((.(((((.((....)).)))))((((((((((.....)))).)).)))).)))) ( -13.30, z-score = -0.09, R) >droSim1.chr3L 14763142 51 + 22553184 ------------UUCAUUCUCUUUUCC----CACUUUCGGUGUCUCGUUUAGAUACCGAAAGGAUGG ------------..............(----((((((((((((((.....))))))))))))..))) ( -17.90, z-score = -4.28, R) >consensus ___UU_UUUUUUUUCAUUCUGUUUUCC____CACUUUUGGUGUCUCAAUCAGAUACCAAAAGGAUGG ......................................((.((((.....)))).)).......... ( -2.43 = -2.60 + 0.17)
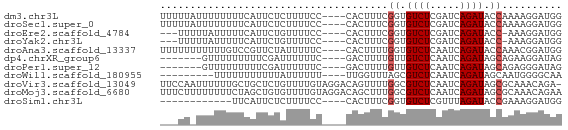
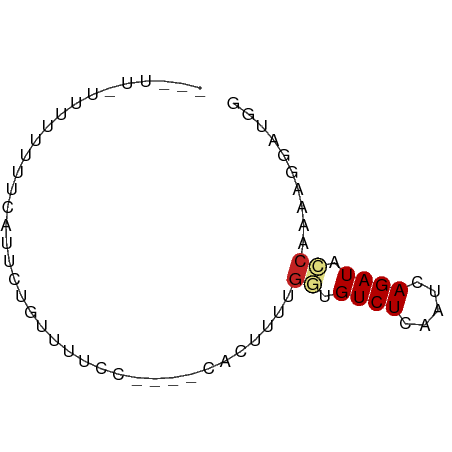

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:28:50 2011