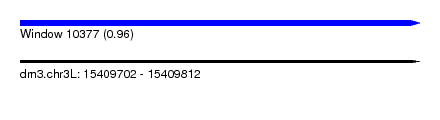
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,409,702 – 15,409,812 |
| Length | 110 |
| Max. P | 0.955377 |

| Location | 15,409,702 – 15,409,812 |
|---|---|
| Length | 110 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.11 |
| Shannon entropy | 0.38473 |
| G+C content | 0.55570 |
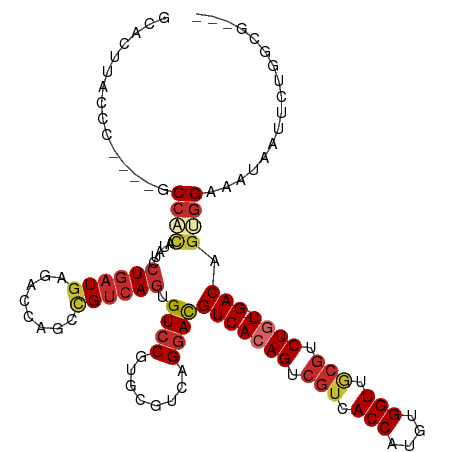
| Mean single sequence MFE | -39.89 |
| Consensus MFE | -24.39 |
| Energy contribution | -25.31 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
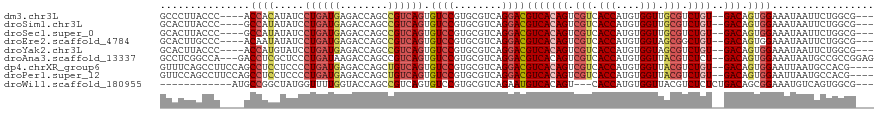
>dm3.chr3L 15409702 110 + 24543557 GCCCUUACCC----ACCACAUAUCCUGAUGAGACCAGCCGUCAGUGUCCGUGCGUCAGGACGUCACAGUCGUCACCAUGUGGUUGCGUCUGU--GACAGUGGAAAUAAUUCUGGCG--- (((.......----.((((...((((((((.((((........).)))....)))))))).(((((((.(((.(((....))).))).))))--))).))))..........))).--- ( -43.07, z-score = -3.31, R) >droSim1.chr3L 14749988 110 + 22553184 GCACUUACCC----GCCAUAUAUCCUGAUGAGACCAGCCGUCAGUGUCCGUGCGUCAGGACGUCACAGUCGUCACCAUGUGGUUGCGUCUGU--GACAGUGGAAAUAAUUCUGGCG--- .........(----((((....((((((((.((((........).)))....)))))))).(((((((.(((.(((....))).))).))))--)))..............)))))--- ( -41.00, z-score = -2.53, R) >droSec1.super_0 7518806 110 + 21120651 GCACUUACCC----GCCAUAUAUCCUGAUGAGACCAGCCGUCAGUGUCCGUGCGUCAGGACGUCACAGUCGUCACCAUGUGGUUGCGUCUGU--GACAGUGGAAAUAAUUCUGGCG--- .........(----((((....((((((((.((((........).)))....)))))))).(((((((.(((.(((....))).))).))))--)))..............)))))--- ( -41.00, z-score = -2.53, R) >droEre2.scaffold_4784 17668689 110 + 25762168 GCACUUGCCC----ACAAUAUAUCCUGAUGAGACCAGCCGUCAGUGUCCGUGCGUCAGGACGUCACAGUCGUCACCAUGUGGUAGCGGCUGU--GACAGUGGAAAUAAUUCUGGCG--- ......((((----((......((((((((.((((........).)))....)))))))).((((((((((..(((....)))..)))))))--))).)))(((....))).))).--- ( -44.40, z-score = -3.14, R) >droYak2.chr3L 17926113 110 + 24197627 GCACUUACCC----ACCAUGUAUCCUGAUGAGACCAGCCGUCAGUGUCCGUGCGUCAGGACGUCACAGUCGUCACCAUGUGGUAGCGUCUGU--GACAGUGGAAAUAAUUCUGGCG--- ((((.(((..----.....)))..((((((........)))))).....))))(((((((.(((((((.((..(((....)))..)).))))--))).((....))..))))))).--- ( -40.00, z-score = -2.11, R) >droAna3.scaffold_13337 5850943 114 - 23293914 GCCUCGGCCA---GACCUCGCUCCCUGAUAAGACCAGCCGUCAGUGUCCGUGCGUCAGGACGUCACAGUCGUCACCAUGUGGUUACGUCUCU--GACAGUGGAAAUAAUGCCGCCGGAG .((.((((..---....(((((.((((((..(((.....))).((......))))))))..((((.((.(((.(((....))).))).)).)--)))))))).......))))..)).. ( -34.82, z-score = 0.04, R) >dp4.chrXR_group6 12968275 113 - 13314419 GUUUCAGCCUUCCAGCCUCCUCCCCUGAUGAGACCAGCUGUCAGUGUCCGUGCGUCAGGACGUCACAGUCGUCACCAUGUGGUUACGUCUGU--GACAGUGGAAUUAAUGCCACG---- ........((..(((((((.((....)).)))....))))..)).((((........))))(((((((.(((.(((....))).))).))))--))).((((........)))).---- ( -38.90, z-score = -2.39, R) >droPer1.super_12 1387178 113 - 2414086 GUUCCAGCCUUCCAGCCUCCUCCCCUGAUGAGACCAGCUGUCAGUGUCCGUGCGUCAGGACGUCACAGUCGUCACCAUGUGGUUACGUCUGU--GACAGUGGAAUUAAUGCCACG---- ((((((..((..(((((((.((....)).)))....))))..)).((((........))))(((((((.(((.(((....))).))).))))--)))..))))))..........---- ( -38.90, z-score = -2.40, R) >droWil1.scaffold_180955 2326636 101 - 2875958 ------------AUGCCGGCUAUGGUUUUGGUACCAGCCGUCAGUGUCCGUGCGUCAGAAUGUCACAGU---CACCAUGUGGUUACGUCUCUCUGACAGCGGAAAUGUCAGUGGCG--- ------------..(((((((..(((......)))))))(.((...(((((..((((((..(.....((---.(((....))).))..)..)))))).)))))..)).)...))).--- ( -36.90, z-score = -1.88, R) >consensus GCACUUACCC____GCCACAUAUCCUGAUGAGACCAGCCGUCAGUGUCCGUGCGUCAGGACGUCACAGUCGUCACCAUGUGGUUGCGUCUGU__GACAGUGGAAAUAAUUCUGGCG___ ...............((((.....((((((........)))))).((((........))))(((((((.(((.(((....))).))).))))..))).))))................. (-24.39 = -25.31 + 0.92)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:28:46 2011