| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,399,251 – 15,399,344 |
| Length | 93 |
| Max. P | 0.672154 |

| Location | 15,399,251 – 15,399,344 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.83 |
| Shannon entropy | 0.39140 |
| G+C content | 0.44765 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -16.60 |
| Energy contribution | -16.61 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.672154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
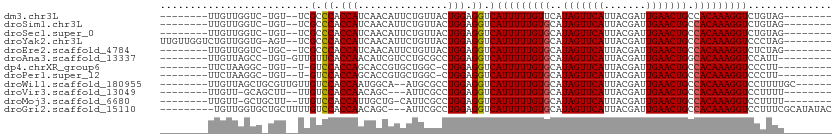
>dm3.chr3L 15399251 93 - 24543557 --------UUGUUGGUC-UGU--UCGCCCACCAUCAACAUUCUGUUACUGGAGGUCAUUUUUGUUCAUAGUUCAUUACGAUUGAACUGCCACAAAGGUCUGUAG-------- --------.(((((((.-.((--......)).)))))))........(((.((..(...(((((...(((((((.......)))))))..))))))..)).)))-------- ( -19.20, z-score = -0.11, R) >droSim1.chr3L 14739420 93 - 22553184 --------UUGUUGGUC-UGU--UCGCCCACCAUCAACAUUCUGUUACUGGAGGUCAUUUUUGUGCAUAGUUCAUUACGAUUGAACUGCCACAAAGGUCUGUAG-------- --------.(((((((.-.((--......)).)))))))........(((.((..(...((((((..(((((((.......))))))).)))))))..)).)))-------- ( -22.00, z-score = -0.60, R) >droSec1.super_0 7508417 93 - 21120651 --------UUGUUGGUC-UGU--UCGCCCACCAUCAACAUUCUGUUACUGGAGGUCAUUUUUGUGCAUAGUUCAUUACGAUUGAACUGCCACAAAGGUCUGUAG-------- --------.(((((((.-.((--......)).)))))))........(((.((..(...((((((..(((((((.......))))))).)))))))..)).)))-------- ( -22.00, z-score = -0.60, R) >droYak2.chr3L 17915199 101 - 24197627 UUGUUGGUCUGUUGGUG-AGU--UCGCCCACCAUCAACAUUCUGUUACUGGAGGUCAUUUUUGUGCAUAGUUCAUUACGAUUGAACUGCCACAAAGGUCCCUAG-------- .(((((((..((.((((-...--.)))).)).)))))))........((((.((..(((((((((..(((((((.......))))))).)))))))))))))))-------- ( -31.10, z-score = -2.36, R) >droEre2.scaffold_4784 17658008 93 - 25762168 --------UUGUUGGUC-UGC--UCGCCCACCAUCAACAUUCUGUUACUGGAGGUCAUUUUUGUGCAUAGUUCAUUACGAUUGAACUGCCACAAAGGUCUCUAG-------- --------(((.((((.-.(.--...)..)))).)))..........((((((..(...((((((..(((((((.......))))))).)))))))..))))))-------- ( -26.00, z-score = -2.26, R) >droAna3.scaffold_13337 5832594 93 + 23293914 --------UUGUUAGCC-UGU-GUUGUUCACCAACAUCGUCCUGCGCCUGGAGGUCAUUUUUGUGCAUAGUUCAUUACGAUUGAACUGGCACAAAGGUCCAUU--------- --------......(((-(((-((((.....)))))).(.(((.(....).))).)....((((((.(((((((.......))))))))))))))))).....--------- ( -27.20, z-score = -1.74, R) >dp4.chrXR_group6 12957671 90 + 13314419 --------UUCUAAGGC-UGU--U-GUCCACCAGCACCGUGCUGGC-CUGGAGGUCAUUUUUGUGCAUAGUUCAUUACGAUUGAACUGCCACAAAGGUCCCUU--------- --------......(((-(..--.-.(((((((((.....))))).-.))))))))(((((((((..(((((((.......))))))).))))))))).....--------- ( -28.50, z-score = -1.74, R) >droPer1.super_12 1376547 90 + 2414086 --------UUCUAAGGC-UGU--U-GUCCACCAGCACCGUGCUGGC-CUGGAGGUCAUUUUUGUGCAUAGUUCAUUACGAUUGAACUGCCACAAAGGUCCCUU--------- --------......(((-(..--.-.(((((((((.....))))).-.))))))))(((((((((..(((((((.......))))))).))))))))).....--------- ( -28.50, z-score = -1.74, R) >droWil1.scaffold_180955 2315865 96 + 2875958 --------UUGUUAGCUGCGUUGUUGUCCACCAAUGGCA--AUGCGCCUGGAGGUCAUUUUUGUGCAUAGUUCAUUACGAUUGAACUGCCACAAAGGUCCUUUUGC------ --------....(((.((((((((..(......)..)))--))))).)))((((..(((((((((..(((((((.......))))))).)))))))))))))....------ ( -29.90, z-score = -1.80, R) >droVir3.scaffold_13049 20178941 90 - 25233164 --------UUGUU-GCAGCUU--UUGUCCACCAACAGC---AUUCGCCUGGAGGUCAUUUUUGUGCAUAGUUCAUUACGAUUGAACUGCCACAAAGGUCCUUUU-------- --------.....-...(((.--(((.....))).)))---........(((((..(((((((((..(((((((.......))))))).)))))))))))))).-------- ( -21.40, z-score = -0.85, R) >droMoj3.scaffold_6680 7880165 92 - 24764193 --------UUGUU-GCUGCUU--UUGUCCACCAUUGCUG-CAUUCGCCUGGAGGUCAUUUUUGUGCAUAGUUCAUUACGAUUGAACUGCCACAAAGGUCCUUUU-------- --------....(-((.((..--.((.....))..)).)-)).......(((((..(((((((((..(((((((.......))))))).)))))))))))))).-------- ( -22.00, z-score = -0.71, R) >droGri2.scaffold_15110 4580307 100 + 24565398 ---------UGUUGGUGCUGCUUUUGUCCACCAACAGC---AUUCGCCUGGAGGUCAUUUUUGUGCAUAGUUCAUUACGAUUGAACUGCCACAAAGGUCCUUUCGCAUAUAC ---------((((((((..((....)).))))))))..---....((..(((((..(((((((((..(((((((.......))))))).)))))))))))))).))...... ( -31.00, z-score = -2.65, R) >consensus ________UUGUUGGUC_UGU__UCGUCCACCAUCAACAUUCUGCGACUGGAGGUCAUUUUUGUGCAUAGUUCAUUACGAUUGAACUGCCACAAAGGUCCCUUG________ .........................(.((.(((...............))).)).)(((((((((..(((((((.......))))))).))))))))).............. (-16.60 = -16.61 + 0.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:28:46 2011