
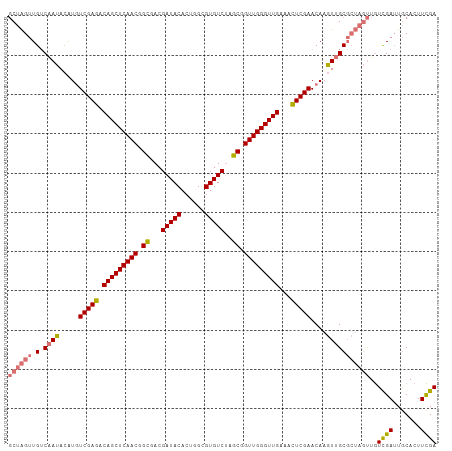
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,290,633 – 15,290,828 |
| Length | 195 |
| Max. P | 0.922722 |

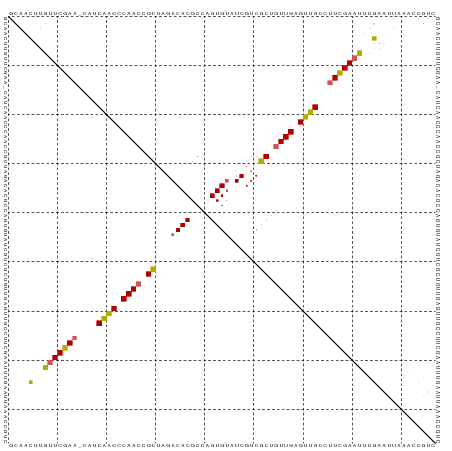
| Location | 15,290,633 – 15,290,742 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.86 |
| Shannon entropy | 0.24562 |
| G+C content | 0.50465 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -31.34 |
| Energy contribution | -31.78 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.814689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15290633 109 - 24543557 GCUAGUUGUCAAUACAUGUCGAGACAGCUCAACGGCGACGAUACACUGGCGUGUCUAGCGGUUGGGUUGAAACUCGAACAAGUUGCGCUAGUUGUCGAUUGCACUUCGA ((((((.(.((((.....(((((.(((((((((.((...(((((......)))))..)).)))))))))...)))))....)))))))))))..((((.......)))) ( -37.90, z-score = -1.50, R) >droSim1.chrX 15891148 109 - 17042790 GCUAGUUGUCAAUACAUGUCGAGACAGCUCAACGGCGACGAUACACUGGCGUGUCUAGCGGUUGGGUUGAAACUCGAACAAGUUGCGCUAGUUGUCGAUUGCACUUCGA ((((((.(.((((.....(((((.(((((((((.((...(((((......)))))..)).)))))))))...)))))....)))))))))))..((((.......)))) ( -37.90, z-score = -1.50, R) >droSec1.super_232 14022 103 - 21018 ACCGGGUGGCCACAAGU-UCGAAACAGCUCAACAGUGACGAUACACUGGCGUGUCAAACGGUUGGGUUGUGGUUCGAACAAGUAGC-----CGAUUGAGUACAGUUUGA (((((.((((.((..((-(((((((((((((((.((...(((((......)))))..)).))))))))))..)))))))..)).))-----)).))).))......... ( -36.60, z-score = -1.98, R) >droYak2.chr3L_random 2168867 109 - 4797643 GCUAGUUGUCAAUACAUGUCGAGACAGCUCAACGGCGACGAUACACUGGCGUGUCUAGCGGUUGGGUUGAAACUCGAACAAGUUGCGCUAGUUGUCGAUUGCACUUCGA ((((((.(.((((.....(((((.(((((((((.((...(((((......)))))..)).)))))))))...)))))....)))))))))))..((((.......)))) ( -37.90, z-score = -1.50, R) >consensus GCUAGUUGUCAAUACAUGUCGAGACAGCUCAACGGCGACGAUACACUGGCGUGUCUAGCGGUUGGGUUGAAACUCGAACAAGUUGCGCUAGUUGUCGAUUGCACUUCGA ((((((.(.((((.....(((((.(((((((((.((...(((((......)))))..)).)))))))))...)))))....)))))))))))..((((.......)))) (-31.34 = -31.78 + 0.44)


| Location | 15,290,742 – 15,290,828 |
|---|---|
| Length | 86 |
| Sequences | 5 |
| Columns | 87 |
| Reading direction | forward |
| Mean pairwise identity | 85.19 |
| Shannon entropy | 0.26415 |
| G+C content | 0.49893 |
| Mean single sequence MFE | -20.62 |
| Consensus MFE | -19.32 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15290742 86 + 24543557 GCAACUUGUUCGAA-CAUCAACCCAACCGCUAGACACGCCAGUGUAUCGUCGCUGUUGAGUUGCCUUCGAAUUUGAAUUAAACCGUC ....(..(((((((-...((((.((((.((...((((....))))......)).)))).))))..)))))))..)............ ( -21.40, z-score = -2.14, R) >droSim1.chrX 15891257 86 + 17042790 GCAACUUGUUCGAA-CAUCAACCCAACCGCUAGACACGCCAGUGUAUCGUCGCUGUUGAGUUGCCUUUGAAUUUGAAUUAAACCGUC ....(..(((((((-...((((.((((.((...((((....))))......)).)))).))))..)))))))..)............ ( -19.30, z-score = -1.45, R) >droSec1.super_232 14130 86 + 21018 GCAAUCUGCUCGAACCAUCGACCCAACCGUUUGACACGCCAGUGUAUCGUCACUGUUGAGUUGC-UUCGAACAUAUGCUCGACCGGC (((...((.(((((....((((.((((.((...((((....))))......)).)))).)))).-))))).))..)))......... ( -18.60, z-score = 0.35, R) >droYak2.chr3L_random 2168976 86 + 4797643 GCAACUUGUUCGAA-CAUCAACCCAACCGCUAGACACGCCAGUGUAUCGUCGCUGUUGAGUUGCCUUCGAAUUUGAAUUAAACCGUC ....(..(((((((-...((((.((((.((...((((....))))......)).)))).))))..)))))))..)............ ( -21.40, z-score = -2.14, R) >droEre2.scaffold_4929 2234929 85 - 26641161 GCAACUUGUUCGA--CAUCAGCCCAACAGCUAGACACGCCAGUGGAUCGUCGCUAUUGAGCUGUACUCGAACUCGAACUUGGCCGUC .......((((((--...((((.(((.(((..(((((....)))..))...))).))).))))...))))))............... ( -22.40, z-score = -0.43, R) >consensus GCAACUUGUUCGAA_CAUCAACCCAACCGCUAGACACGCCAGUGUAUCGUCGCUGUUGAGUUGCCUUCGAAUUUGAAUUAAACCGUC ....(..(((((((....((((.((((.((...((((....))))......)).)))).))))..)))))))..)............ (-19.32 = -18.60 + -0.72)


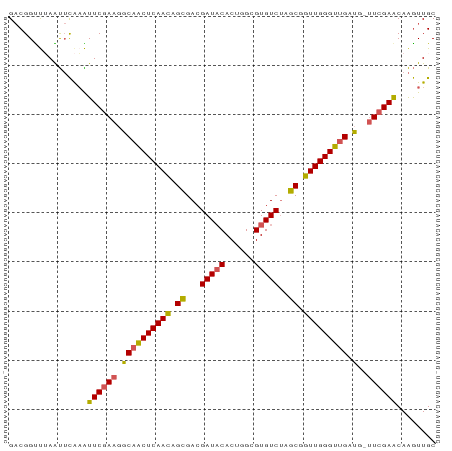
| Location | 15,290,742 – 15,290,828 |
|---|---|
| Length | 86 |
| Sequences | 5 |
| Columns | 87 |
| Reading direction | reverse |
| Mean pairwise identity | 85.19 |
| Shannon entropy | 0.26415 |
| G+C content | 0.49893 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.22 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.922722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15290742 86 - 24543557 GACGGUUUAAUUCAAAUUCGAAGGCAACUCAACAGCGACGAUACACUGGCGUGUCUAGCGGUUGGGUUGAUG-UUCGAACAAGUUGC (((((......))...((((((.((((((((((.((...(((((......)))))..)).))))))))).).-))))))...))).. ( -27.50, z-score = -2.40, R) >droSim1.chrX 15891257 86 - 17042790 GACGGUUUAAUUCAAAUUCAAAGGCAACUCAACAGCGACGAUACACUGGCGUGUCUAGCGGUUGGGUUGAUG-UUCGAACAAGUUGC (((((......))...(((.((.((((((((((.((...(((((......)))))..)).))))))))).).-)).)))...))).. ( -21.60, z-score = -0.68, R) >droSec1.super_232 14130 86 - 21018 GCCGGUCGAGCAUAUGUUCGAA-GCAACUCAACAGUGACGAUACACUGGCGUGUCAAACGGUUGGGUCGAUGGUUCGAGCAGAUUGC .........(((..((((((((-.(((((((((.((...(((((......)))))..)).)))))))...)).))))))))...))) ( -30.00, z-score = -1.67, R) >droYak2.chr3L_random 2168976 86 - 4797643 GACGGUUUAAUUCAAAUUCGAAGGCAACUCAACAGCGACGAUACACUGGCGUGUCUAGCGGUUGGGUUGAUG-UUCGAACAAGUUGC (((((......))...((((((.((((((((((.((...(((((......)))))..)).))))))))).).-))))))...))).. ( -27.50, z-score = -2.40, R) >droEre2.scaffold_4929 2234929 85 + 26641161 GACGGCCAAGUUCGAGUUCGAGUACAGCUCAAUAGCGACGAUCCACUGGCGUGUCUAGCUGUUGGGCUGAUG--UCGAACAAGUUGC ..(((......))).((((((...((((((((((((((((..((...))..))))..))))))))))))...--))))))....... ( -31.00, z-score = -1.78, R) >consensus GACGGUUUAAUUCAAAUUCGAAGGCAACUCAACAGCGACGAUACACUGGCGUGUCUAGCGGUUGGGUUGAUG_UUCGAACAAGUUGC ................((((((..(((((((((.((...(((((......)))))..)).)))))))))....))))))........ (-23.38 = -23.22 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:28:29 2011