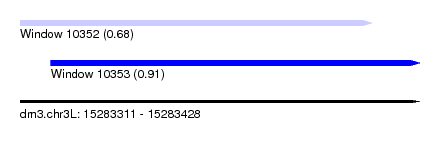
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,283,311 – 15,283,428 |
| Length | 117 |
| Max. P | 0.907089 |

| Location | 15,283,311 – 15,283,414 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 69.13 |
| Shannon entropy | 0.61924 |
| G+C content | 0.42161 |
| Mean single sequence MFE | -23.34 |
| Consensus MFE | -5.65 |
| Energy contribution | -5.42 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15283311 103 + 24543557 -----AAGUUAACAAAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU--UGCUGGCUC-----AUCCCGCAUCUUU---UCUUGCAC-UCGCUUACCGCAUUUCUCGCUG -----...(((((((......)))))))....((((((((..(((...(((((...--(((.((...-----..)).)))...))---)))..)))-....)))))))).......... ( -26.30, z-score = -3.24, R) >droSim1.chr3L 14623952 103 + 22553184 -----AAGUUAACAAAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU--UGCUGGCUC-----AUCCCGCAUCUUU---UCUUGCCC-GCGCUUACCGCAUUUCUCGCUG -----...(((((((......)))))))....((((((((.((((...(((((...--(((.((...-----..)).)))...))---)))....)-))).)))))))).......... ( -27.10, z-score = -3.07, R) >droSec1.super_0 7395237 103 + 21120651 -----AAGUUAACAAAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU--UGCUGGCUC-----AUCCCGCAUCUUU---UCUUGCCC-GCGCUUACCGCAUUUCUCGCUG -----...(((((((......)))))))....((((((((.((((...(((((...--(((.((...-----..)).)))...))---)))....)-))).)))))))).......... ( -27.10, z-score = -3.07, R) >droYak2.chr3L 17795265 101 + 24197627 -----AAGUUAACAAAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU--UGCCGGCUC-----AUCCCGCAUCUUU---UCUUGCC--UCGCUUACCGCAUU-CUCACUG -----...(((((((......)))))))....((((((((..((((..(((((...--(((.((...-----..)).)))...))---)))....--))))))))))))..-....... ( -25.50, z-score = -3.61, R) >droEre2.scaffold_4784 17544047 103 + 25762168 -----AAGUUAGCAAAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU--UGCUGGCUC-----AUCCCGCAUCUUU---UCUUGCCC-UCGCUUACCGCAUUUCUCACUG -----...(((((((......)))))))....((((((((..((((..(((((...--(((.((...-----..)).)))...))---))).....-)))))))))))).......... ( -24.80, z-score = -2.72, R) >droAna3.scaffold_13337 5580605 105 - 23293914 -----AAGUUAACAAAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU--UGCUGCCUCA----UUCCCCCAUCUGC---UCUUCUCUAGCAUUUCCCGCAUUUCGCGCUG -----.(((((((((......))))))((((.(((((.((((((....((((....--.((.......----...........))---..))))...)))))).))))).)).))))). ( -19.27, z-score = -1.64, R) >dp4.chrXR_group6 12845972 112 - 13314419 -----AAGUCAACAAAAAACCUUGUUAGUGAUUGCGGUAAAUGAGAUUAGAAACUUCUUGCAGGCUCCACCUACCCCCCACCCACCUCUCGUGGCC-UCGUUUACCGCUCUUCUCGCA- -----.....(((((......))))).((((..((((((((((((................(((.....))).........((((.....)))).)-))))))))))).....)))).- ( -29.70, z-score = -3.44, R) >droPer1.super_12 1265714 112 - 2414086 -----AAGUCAACAAAAAACCUUGUUAGUGAUUGCGGUAAAUGAGAUUAGAAACUUCUUGCUGGCUCCACCUGCCCCCCACCCUCCACUCGUGGCC-UCGUUUACCGCUCUUCUCGCA- -----.....(((((......))))).((((..((((((((((((.................(((.......)))..((((.........)))).)-))))))))))).....)))).- ( -31.10, z-score = -3.64, R) >droWil1.scaffold_180955 685730 98 + 2875958 CUAUGGAAGGAACAAAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUUUUACCACAUUUCGCUGGUCUCCUUCCAGC---UACGGGUUUUUGC------------------ ...((((((((((((......))))((((((.((.((((((.((........)).)))))).)).)).))))...)))))))).(---....)........------------------ ( -24.30, z-score = -1.52, R) >droVir3.scaffold_13049 20067715 79 + 25233164 -----CAGUCAACAAAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAAGUU--UCGUGGAACGUUCUUCUCCUUAGCAAUG--------------------------------- -----................(((((((((((..((.....))..))))(((.(((--(....)))).)))......)))))))..--------------------------------- ( -12.30, z-score = -0.14, R) >droGri2.scaffold_15110 4469077 79 - 24565398 -----CAGUCAACAAAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU--UCUUGCAACAUCCUCCUCCUUAGCAAUG--------------------------------- -----.....(((((......)))))....(((((((...((((....(((.....--)))....))))...))......))))).--------------------------------- ( -9.30, z-score = 0.14, R) >consensus _____AAGUUAACAAAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU__UGCUGGCUC_____AUCCCGCAUCUUU___UCUUGCCC_UCGCUUACCGCAUUUCUCGCUG ..........(((((......)))))..((((..((.....))..))))...................................................................... ( -5.65 = -5.42 + -0.23)
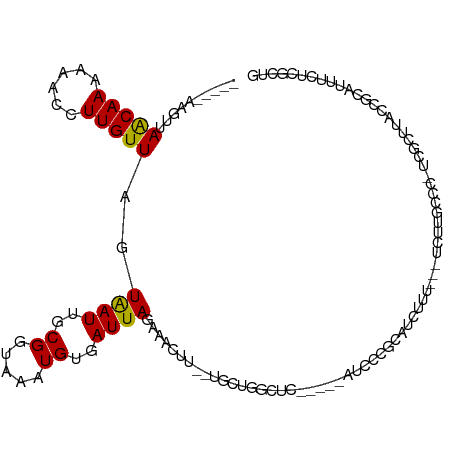
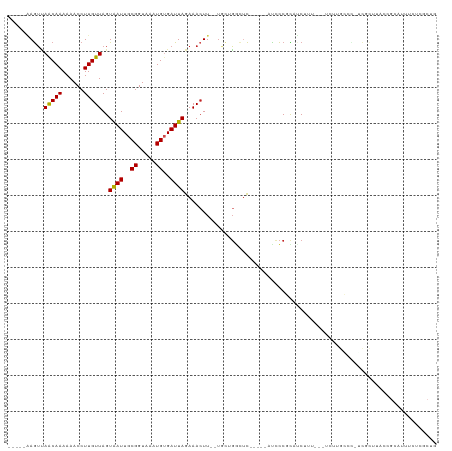
| Location | 15,283,320 – 15,283,428 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 66.92 |
| Shannon entropy | 0.64178 |
| G+C content | 0.42264 |
| Mean single sequence MFE | -22.04 |
| Consensus MFE | -5.25 |
| Energy contribution | -5.19 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15283320 108 + 24543557 AAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU--UGCUGGCUC---------AUCCCGCAUCUUU---UCUUGCAC-UCGCUUACCGCAUUUCUCGCUGUUUUCCCCUAUUUG .(((((...((.((.((.((((((((..(((...(((((...--(((.((...---------..)).)))...))---)))..)))-....)))))))).)))).)).))))).......... ( -26.60, z-score = -3.98, R) >droSim1.chr3L 14623961 108 + 22553184 AAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU--UGCUGGCUC---------AUCCCGCAUCUUU---UCUUGCCC-GCGCUUACCGCAUUUCUCGCUGUUUUCCCCUAUUUG .(((((...((.((.((.((((((((.((((...(((((...--(((.((...---------..)).)))...))---)))....)-))).)))))))).)))).)).))))).......... ( -27.40, z-score = -3.90, R) >droSec1.super_0 7395246 108 + 21120651 AAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU--UGCUGGCUC---------AUCCCGCAUCUUU---UCUUGCCC-GCGCUUACCGCAUUUCUCGCUGUUUUCCCCUAUUUG .(((((...((.((.((.((((((((.((((...(((((...--(((.((...---------..)).)))...))---)))....)-))).)))))))).)))).)).))))).......... ( -27.40, z-score = -3.90, R) >droYak2.chr3L 17795274 106 + 24197627 AAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU--UGCCGGCUC---------AUCCCGCAUCUUU---UCUUGCC--UCGCUUACCGCAUU-CUCACUGUUUUCCCCUAUUUG .(((((...((.((....((((((((..((((..(((((...--(((.((...---------..)).)))...))---)))....--))))))))))))..-)).)).))))).......... ( -24.90, z-score = -4.04, R) >droEre2.scaffold_4784 17544056 108 + 25762168 AAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU--UGCUGGCUC---------AUCCCGCAUCUUU---UCUUGCCC-UCGCUUACCGCAUUUCUCACUGUUUUCCCCUAUUUG .(((((...((.((.((.((((((((..((((..(((((...--(((.((...---------..)).)))...))---))).....-)))))))))))).)))).)).))))).......... ( -24.80, z-score = -4.05, R) >droAna3.scaffold_13337 5580614 110 - 23293914 AAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU--UGCUGCCUCA--------UUCCCCCAUCUGC---UCUUCUCUAGCAUUUCCCGCAUUUCGCGCUGUUUUCCCCUAUUUG .(((((..(((.((....(((((.((((((....((((....--.((.......--------...........))---..))))...)))))).))))).)).)))..))))).......... ( -17.07, z-score = -1.56, R) >dp4.chrXR_group6 12845981 117 - 13314419 AAAAACCUUGUUAGUGAUUGCGGUAAAUGAGAUUAGAAACUUCUUGCAGGCUCCACCU----ACCCCCCACCCACCUCUCGUGGCC-UCGUUUACCGCUCUUCUCGCAAAUU-CCCCUGUUUG .......((((.((.....((((((((((((................(((.....)))----.........((((.....)))).)-)))))))))))....)).))))...-.......... ( -28.30, z-score = -3.45, R) >droPer1.super_12 1265723 117 - 2414086 AAAAACCUUGUUAGUGAUUGCGGUAAAUGAGAUUAGAAACUUCUUGCUGGCUCCACCU----GCCCCCCACCCUCCACUCGUGGCC-UCGUUUACCGCUCUUCUCGCAAAUU-CCCCUGUUUG .......((((.((.....((((((((((((.................(((.......----)))..((((.........)))).)-)))))))))))....)).))))...-.......... ( -29.70, z-score = -3.88, R) >droWil1.scaffold_180955 685744 95 + 2875958 AAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU-UUACCACAUUUCGCUGGUCUCCUUCCAGCUACGGGUUUUUGCUACUAUAUUUG--------------------------- ((((((((..........((.((((((.((........)).)-))))).))....(((((.......)))))...)))))))).............--------------------------- ( -23.00, z-score = -2.60, R) >droVir3.scaffold_13049 20067724 84 + 25233164 AAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAAGUU--UCGUGGAACGUUCU----UCUCCUUAGCAAU---GCUUUCCC-CUAUUUG----------------------------- .......(((((((((((..((.....))..))))(((.(((--(....)))).))).----.....))))))).---........-.......----------------------------- ( -12.30, z-score = -0.25, R) >droMoj3.scaffold_6680 7771149 83 + 24764193 -AAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU--UGGUGCAACGUCCU----UCUCCUUGGCAAU---GCUUUCCC-CCAUUUG----------------------------- -...(((..(((..((((..((.....))..))))..)))..--.)))(((..(.((.----.......)))..)---))......-.......----------------------------- ( -13.60, z-score = -0.11, R) >droGri2.scaffold_15110 4469086 84 - 24565398 AAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU--UCUUGCAACAUCCU----CCUCCUUAGCAAU---GCUUUCCC-CUAUUUG----------------------------- .........(..((((.((((((...((((....(((.....--)))....))))...----))......)))))---)))..)..-.......----------------------------- ( -9.40, z-score = -0.31, R) >consensus AAAAACCUUGUUAGUAAUUGCGGUAAAUGUGAUUAGAAACUU__UGCUGGCUC_________ACCCCGCAUCUUU___UCUUGCCC_UCGUUUACCGCAUUUCUCGCUGUUU_CCCCUAUUUG .........(((..((((..((.....))..))))..)))................................................................................... ( -5.25 = -5.19 + -0.05)

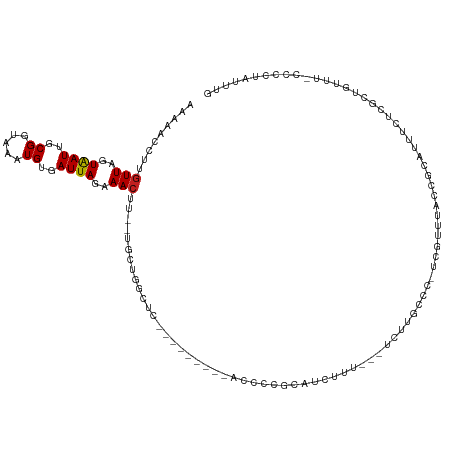
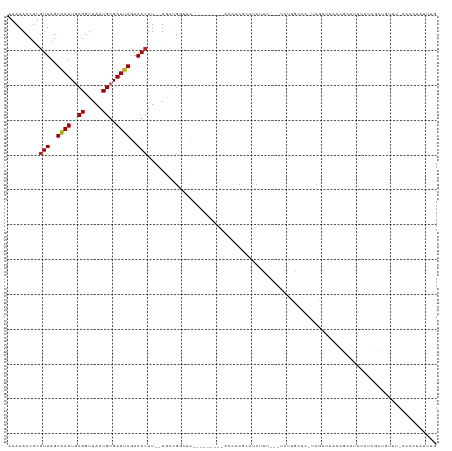
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:28:26 2011