| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,275,072 – 15,275,172 |
| Length | 100 |
| Max. P | 0.622985 |

| Location | 15,275,072 – 15,275,172 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 65.42 |
| Shannon entropy | 0.69970 |
| G+C content | 0.50631 |
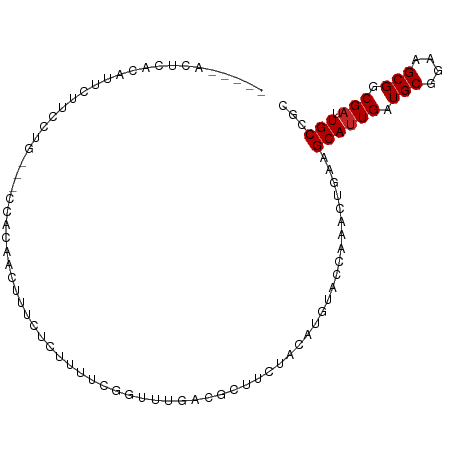
| Mean single sequence MFE | -21.58 |
| Consensus MFE | -9.41 |
| Energy contribution | -9.51 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.78 |
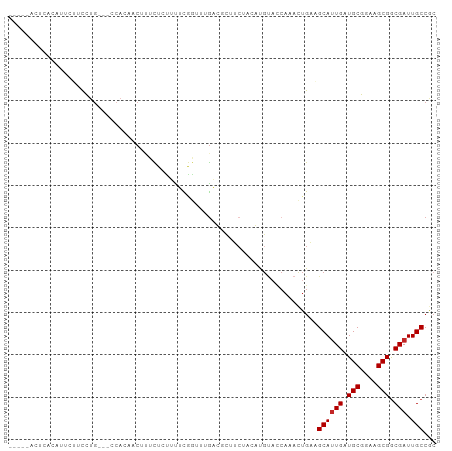
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
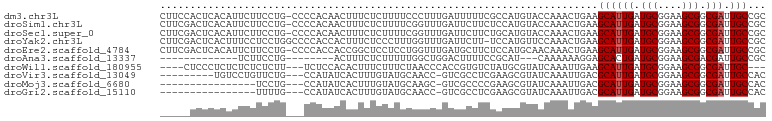
>dm3.chr3L 15275072 100 + 24543557 CUUCCACUCACAUUCUUCCUG-CCCCACAACUUUCUCUUUUCCCUUUGAUUUUUCGCCAUGUACCAAACUGAAGCAUUGAUGCGGAAGCGGCGAUUGCCGC .((((................-.................(((..((((................))))..)))(((....)))))))(((((....))))) ( -15.09, z-score = 0.18, R) >droSim1.chr3L 14615303 100 + 22553184 CUUCGACUCACAUUCUUCCUG-CCCCACAACUUUCUCUUUUCGGUUUGAUUCUUCUCCAUGUACCAAACUGAAGCAUUGAUGCGGAAGCGGCGAUUGCCGC ...............((((..-.................(((((((((...(........)...)))))))))(((....)))))))(((((....))))) ( -22.70, z-score = -1.46, R) >droSec1.super_0 7387023 100 + 21120651 CUUCGACUCACAUUCUUCCUG-CCCCACAACUUUCUCUUUUCGGUUUGAUUCUUCUGCAUGUACCAAACUGAAGCAUUGAUGCGGAAGCGGCGAUUGCCGC ...............((((..-.................(((((((((...(........)...)))))))))(((....)))))))(((((....))))) ( -22.70, z-score = -1.04, R) >droYak2.chr3L 17786798 100 + 24197627 CUUCGACUCACUUUCCUCCUGGCCCCACCACUUUCUCCCUUUGGUUUGAUUCUU-UCCAUGUUCCAAACUGAAGCAUUGAUGCGGAAGCGGCGAUUGCCGC .((((..(((.........(((.....)))........(((..(((((...(..-.....)...)))))..)))...)))..)))).(((((....))))) ( -21.60, z-score = -0.52, R) >droEre2.scaffold_4784 17535517 100 + 25762168 CUUCGACUCACAUUCUUCCUG-CCCCACCACCGGCUCCUCCUGGUUUGAUGCUUCUCCAUGCAACAAACUGAAGCAUUGAUGCGGAAGCGGCGAUUGCCGC .((((..(((.((.((((..(-((........))).......((((((.(((........))).)))))))))).)))))..)))).(((((....))))) ( -28.20, z-score = -1.77, R) >droAna3.scaffold_13337 5572882 77 - 23293914 -------------UCUUCCUG--------ACUUUCUCUUUUUGGCUGGACUUUUCCGCAU---CAAAAAAGGAGCACUGAUGCGGAAGCGACGAUUGCCGC -------------........--------.............(((......(((((((((---((............)))))))))))........))).. ( -19.04, z-score = -0.39, R) >droWil1.scaffold_180955 2179825 91 - 2875958 ----CUCCCUCUCUCUCUCUU---UCUCCACACUUUCUUUCUAACCCACCGUGUCUAUGCGUAUCAAAUUGAAGCAUUGAUGCGGAAGCGGCGAUUGC--- ----.................---........................(((((((.((((...((.....)).)))).))))))).............--- ( -15.10, z-score = -0.36, R) >droVir3.scaffold_13049 20060170 88 + 25233164 ---------UGUCCUGUUCUG---CCAUAUCACUUUGUAUGCAACC-GUCGCCUCGAAGCGUAUCAAAUUGACGCAUUGAUGCGGAAGCGGCGAUUGCCAC ---------..........((---(.((((......)))))))...-((((((((((.((((.........)))).)))).((....))))))))...... ( -23.40, z-score = -0.26, R) >droMoj3.scaffold_6680 7763309 81 + 24764193 ----------------UCCUG---CCAUAUCACUUUGUAUGCAAGC-GUCGCCCCGAAGCGUAUCAAAUUGACGCAUUGAUGCGGAAGCGGCGAUUGCCAC ----------------...((---(.((((......))))))).((-((((((.(((.((((.........)))).)))..((....))))))).)))... ( -23.90, z-score = -0.86, R) >droGri2.scaffold_15110 4460955 81 - 24565398 ----------------UUUUG---CCAUAUCACUUUGUAUGCAACC-GUCGCCUCGAAGCGUAUCAAAUUGACGCAUUGAUGCGGAAGCGGCGAUUGCCAC ----------------..(((---(.((((......))))))))..-((((((((((.((((.........)))).)))).((....))))))))...... ( -24.10, z-score = -1.30, R) >consensus _____ACUCACAUUCUUCCUG___CCACAACUUUCUCUUUUCGGUUUGACGCUUCUACAUGUACCAAACUGAAGCAUUGAUGCGGAAGCGGCGAUUGCCGC .........................................................................((((((.(((....))).))).)))... ( -9.41 = -9.51 + 0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:28:23 2011