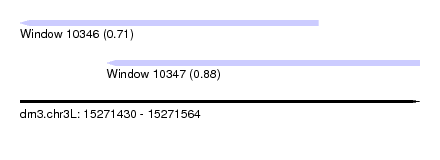
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,271,430 – 15,271,564 |
| Length | 134 |
| Max. P | 0.877074 |

| Location | 15,271,430 – 15,271,530 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 74.64 |
| Shannon entropy | 0.47194 |
| G+C content | 0.50198 |
| Mean single sequence MFE | -17.29 |
| Consensus MFE | -11.34 |
| Energy contribution | -11.73 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
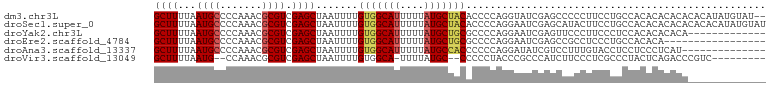
>dm3.chr3L 15271430 100 - 24543557 GCUUUUAAUGCCCCAAACGCGUCGAGCUAAUUUUGUGGCAUUUUUAUGCUACACCCCAGGUAUCGAGCCCCCUUCCUGCCACACACACACACAUAUGUAU-- ((((...((((.......)))).))))......((((((((....)))))))).....((((..(((....)))..))))....................-- ( -19.20, z-score = -1.25, R) >droSec1.super_0 7383472 102 - 21120651 GCUUUUAAUGCCCCAAACGCGUCGAGCUAAUUUUGUGGCAUUUUUAUGCUACACCCCAGGAAUCGAGCAUACUUCCUGCCACACACACACACACAUAUGUAU ((((...((((.......)))).))))......(((((((....((((((....(....).....)))))).....)))))))................... ( -21.00, z-score = -1.79, R) >droYak2.chr3L 17782996 89 - 24197627 GCUUUUAAUGCCCCAAACGCGUCGAGCUAAUUUUGUGGCAUUUUUAUGCUGCGCCCCAGGAAUCGAGUUCCCUUCCCUCCACACACACA------------- ((((...((((.......)))).)))).......(..((((....))))..)......(((...(((....)))...))).........------------- ( -16.00, z-score = -0.25, R) >droEre2.scaffold_4784 17531992 85 - 25762168 GCUUUUAAUGCCCCAAACGCGUCGAGCUAAUUUUGUGGCAUUUUUAUGCUGCGCCCCAGGAAUCGAGCCGCCUCCCUGCCACACA----------------- ((((...((((.......)))).))))......(((((((.......((.((..............)).)).....)))))))..----------------- ( -17.84, z-score = 0.32, R) >droAna3.scaffold_13337 5569612 88 + 23293914 GCUUUUAAUGCCCCAAACGCGUCGAGCUAAUUUUGUGGCAUUUUUAUGCCACCCCCCAGGAUAUCGUCCUUUGUACCUCCUCCCUCAU-------------- ((((...((((.......)))).)))).......(((((((....))))))).....(((((...)))))..................-------------- ( -18.20, z-score = -2.79, R) >droVir3.scaffold_13049 20055835 88 - 25233164 GCUUUUAAUG--CCAAACGCGUCGAGCUAAUUUUGUGGCA-UUUUAUGC--CCCCCUACCCGCCCAUCUUCCCUCGCCCUACUCAGACCCGUC--------- ((.......)--)...(((.((((((..........((((-(...))))--).........((............))....))).))).))).--------- ( -11.50, z-score = -0.47, R) >consensus GCUUUUAAUGCCCCAAACGCGUCGAGCUAAUUUUGUGGCAUUUUUAUGCUACACCCCAGGAAUCGAGCCUCCUUCCCGCCACACACACA_____________ ((((...((((.......)))).)))).......(((((((....))))))).................................................. (-11.34 = -11.73 + 0.39)
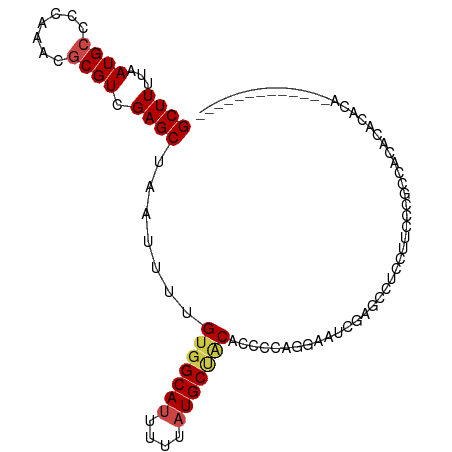

| Location | 15,271,459 – 15,271,564 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.80 |
| Shannon entropy | 0.52725 |
| G+C content | 0.48400 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -11.68 |
| Energy contribution | -12.52 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
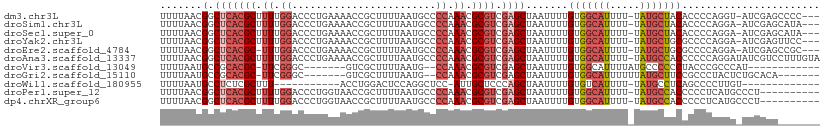
>dm3.chr3L 15271459 105 - 24543557 UUUUAACGGCUCACGCUUUUGGACCCUGAAAACCGCUUUUAAUGCCCCAAACGCGUCGAGCUAAUUUUGUGGCAUUUU-UAUGCUACACCCCAGGU-AUCGAGCCCC--- .......(((((((((.(((((.(..(((((.....)))))..)..))))).)))).(((((.....((((((((...-.)))))))).....)))-.)))))))..--- ( -30.20, z-score = -3.07, R) >droSim1.chr3L 14611853 105 - 22553184 UUUUAACGGCUCACGCUUUUGGACCCUGAAAACCGCUUUUAAUGCCCCAAACGCGUCGAGCUAAUUUUGUGGCAUUUU-UAUGCUACACCCCAGGA-AUCGAGCAUA--- .......(((((((((.(((((.(..(((((.....)))))..)..))))).)))).))))).....((((((((...-.))))))))........-..........--- ( -27.70, z-score = -2.22, R) >droSec1.super_0 7383503 105 - 21120651 UUUUAACGGCUCACGCUUUUGGACCCUGAAAACCGCUUUUAAUGCCCCAAACGCGUCGAGCUAAUUUUGUGGCAUUUU-UAUGCUACACCCCAGGA-AUCGAGCAUA--- .......(((((((((.(((((.(..(((((.....)))))..)..))))).)))).))))).....((((((((...-.))))))))........-..........--- ( -27.70, z-score = -2.22, R) >droYak2.chr3L 17783014 105 - 24197627 UUUUAACGGCUCACGCUUUUGGACCCUGAAAACCGCUUUUAAUGCCCCAAACGCGUCGAGCUAAUUUUGUGGCAUUUU-UAUGCUGCGCCCCAGGA-AUCGAGUUCC--- .......(((((((((.(((((.(..(((((.....)))))..)..))))).)))).)))))......(..((((...-.))))..)......(((-((...)))))--- ( -28.20, z-score = -1.46, R) >droEre2.scaffold_4784 17532006 104 - 25762168 UUUUAACGGCUCACGC-UUUGGACCCUGAAAACCGCUUUUAAUGCCCCAAACGCGUCGAGCUAAUUUUGUGGCAUUUU-UAUGCUGCGCCCCAGGA-AUCGAGCCGC--- ......((((((((((-(((((.(..(((((.....)))))..)..))))).)))).((........((..((((...-.))))..))........-.)))))))).--- ( -28.83, z-score = -1.02, R) >droAna3.scaffold_13337 5569625 109 + 23293914 UUUUAACGGCUCACGCUUUUGGACCCUGAAAACCGCUUUUAAUGCCCCAAACGCGUCGAGCUAAUUUUGUGGCAUUUU-UAUGCCACCCCCCAGGAUAUCGUCCUUUGUA .......(((((((((.(((((.(..(((((.....)))))..)..))))).)))).)))))......(((((((...-.))))))).....(((((...)))))..... ( -31.40, z-score = -3.58, R) >droVir3.scaffold_13049 20055861 88 - 25233164 UUUUAAUGCCGCACGC-UUCGGGC-------GUCGCUUUUAAUG--CCAAACGCGUCGAGCUAAUUUUGUGGCAUUUUAUGCCCCCCUACCCGCCCAU------------ ....(((((((((.((-(.(((((-------((.((.......)--)...)))).))))))......)))))))))......................------------ ( -21.60, z-score = -0.36, R) >droGri2.scaffold_15110 4456982 93 + 24565398 UUUUAAUGCCGCACGC-UUCGGGC-------GUCGCUUUUAAUG--CCAAACGCGUCGAGCUAAUUUUGUGGCAUUUUUUAUGCUUCCGCCCUACUCUGCACA------- ..........(((...-...((((-------(..((....((((--(((...((.....))........)))))))......))...))))).....)))...------- ( -22.60, z-score = -0.17, R) >droWil1.scaffold_180955 2175639 84 + 2875958 UUUUAAUGCCUCUCGCUUU-----------ACCUGGACUCCAGGCUCC-AUUGCUCCCAGCUAAUUUUGUGUCAUUUU-UAUGCCUCAGCCCCUUGU------------- ..............(((..-----------.(((((...)))))....-...((.....)).................-........))).......------------- ( -8.60, z-score = 1.58, R) >droPer1.super_12 1254273 99 + 2414086 UUUUAACGGCUCACGCUUUUGGACCCUGGUAACCGCUUUUAAUGCCCCAAACGCGUCGAGCUAAUUUUGUGGCAUUUU-UAUGCCACCCCCUCAUGCCCU---------- .......(((((((((.(((((((....))....((.......)).))))).)))).)))))......(((((((...-.))))))).............---------- ( -30.60, z-score = -3.99, R) >dp4.chrXR_group6 12834497 99 + 13314419 UUUUAACGGCUCACGCUUUUGGACCCUGGUAACCGCUUUUAAUGCCCCAAACGCGUCGAGCUAAUUUUGUGGCAUUUU-UAUGCCACCCCCUCAUGCCCU---------- .......(((((((((.(((((((....))....((.......)).))))).)))).)))))......(((((((...-.))))))).............---------- ( -30.60, z-score = -3.99, R) >consensus UUUUAACGGCUCACGCUUUUGGACCCUGAAAACCGCUUUUAAUGCCCCAAACGCGUCGAGCUAAUUUUGUGGCAUUUU_UAUGCUACACCCCAGGA_AUCGAG_______ .......(.(((((((.((.((........................)).)).)))).)))).......(((((((.....)))))))....................... (-11.68 = -12.52 + 0.84)
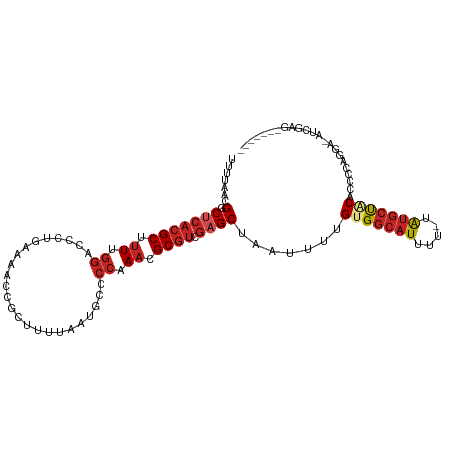
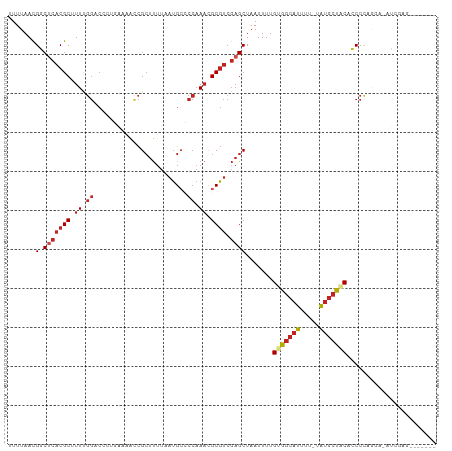
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:28:22 2011