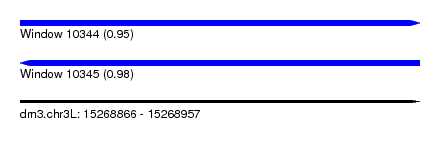
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,268,866 – 15,268,957 |
| Length | 91 |
| Max. P | 0.983637 |

| Location | 15,268,866 – 15,268,957 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.54 |
| Shannon entropy | 0.40400 |
| G+C content | 0.49912 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -19.14 |
| Energy contribution | -19.05 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15268866 91 + 24543557 GCACAAACAACAACAAUAAAUGACAAUAAGAGCGCAGUUCAAGUGUCAGCCGUG--GCCUCUUAUUGUCACGCAUUGUCGUCUGCCCCACUGC------------------ (((....((((.(((((...(((((((((((((((.(((........))).)))--..))))))))))))...))))).)).))......)))------------------ ( -22.40, z-score = -1.29, R) >droAna3.scaffold_13337 5567144 104 - 23293914 ---AUAGGGACAACAAUAAAUGACAAUAAGAGCGCAGUUCAAGUGUCAGUCGUG--GCCUCUUAUUGUCACGCAUUGUCGUCUGCCCCC--GCCCAAGCCCCUGCCACUGC ---.(((((((.(((((...((((((((((((.((....(........).....--))))))))))))))...))))).))).......--((....)).))))....... ( -28.50, z-score = -1.49, R) >droEre2.scaffold_4784 17529495 92 + 25762168 GCACAAACAACAACAAUAAAUGACAAUAAGAGCGCAGUUCAAGUGUCAGCCGUG--GCCUCUUAUUGUCACGCAUUGUCGUCUGCCUCACUGCC----------------- (((....((((.(((((...(((((((((((((((.(((........))).)))--..))))))))))))...))))).)).))......))).----------------- ( -23.20, z-score = -1.49, R) >droYak2.chr3L 17780475 92 + 24197627 GCACAAACAACAACAAUAAAUGACAAUAAGAGCGCAGUUCAAGUGUCAGCCGUG--GCCUCUUAUUGUCACGCAUUGUCGUCUGCCCCACUACC----------------- (((......((.(((((...(((((((((((((((.(((........))).)))--..))))))))))))...))))).)).))).........----------------- ( -22.10, z-score = -1.91, R) >droSec1.super_0 7380903 91 + 21120651 GCACAAACAACAACAAUAAAUGACAAUAAGAGCGCAGUUCAAGUGUCAGCCGUG--GCCUCUUAUUGUCACGCAUUGUCGUCUGCCCCACUGC------------------ (((....((((.(((((...(((((((((((((((.(((........))).)))--..))))))))))))...))))).)).))......)))------------------ ( -22.40, z-score = -1.29, R) >droSim1.chr3L 14609223 91 + 22553184 GCGCAAACAACAACAAUAAAUGACAAUAAGAGCGCAGUUCAAGUGUCAGCCGUG--GCCUCUUAUUGUCACGCAUUGUCGUCUGCCCCACUGC------------------ ..(((.((....(((((...(((((((((((((((.(((........))).)))--..))))))))))))...))))).)).)))........------------------ ( -23.90, z-score = -1.41, R) >droGri2.scaffold_15110 4454391 83 - 24565398 --------AACAACAAUAAAUGACAAUAAGAGCGCAGUUCAAGUGUCAGCCGUG--GCCUCUUAUUGUCACGUGUUGUCCUACCCCCUG--GCCC---------------- --------....((((((..(((((((((((((((.(((........))).)))--..))))))))))))..))))))((........)--)...---------------- ( -24.30, z-score = -3.29, R) >droWil1.scaffold_180955 2172800 95 - 2875958 ------AAAGCAACAAUAAAUGGCAAUAAGAGCGCAGUUCAAGUGUCAGCCGUGUGGCCUCUUAUUGUCACGCGUUGUCGUCUGCCCCACCUUCAGUUACC---------- ------...((((((((...(((((((((((((((.......)))...(((....)))))))))))))))...)))))....)))................---------- ( -23.70, z-score = -1.32, R) >dp4.chrXR_group6 12831228 108 - 13314419 -CAACAGCAACAAAACUAAAUGACAAUAGGAGCGCAGUUCAAGUGUCAGCCGUG--GCCUCUUAUUGUCACGCAUUGUCGUCUGCCCCCAAGUCCCAGACCGCACCAGUCC -.....((............(((((((((((((((.(((........))).)))--..)))))))))))).........(((((...........))))).))........ ( -24.60, z-score = -1.05, R) >droPer1.super_12 1250984 108 - 2414086 -CAACAGCAACAAAACUAAAUGACAAUAGGAGCGCAGUUCAAGUGUCAGCCGUG--GCCUCUUAUUGUCACGCAUUGUCGUCUGCCCCCAAGUCCCAGACCGCACCAGUCC -.....((............(((((((((((((((.(((........))).)))--..)))))))))))).........(((((...........))))).))........ ( -24.60, z-score = -1.05, R) >consensus _CACAAACAACAACAAUAAAUGACAAUAAGAGCGCAGUUCAAGUGUCAGCCGUG__GCCUCUUAUUGUCACGCAUUGUCGUCUGCCCCACUGCC_________________ .........((.(((((...(((((((((((((((.(((........))).)))....))))))))))))...))))).)).............................. (-19.14 = -19.05 + -0.09)

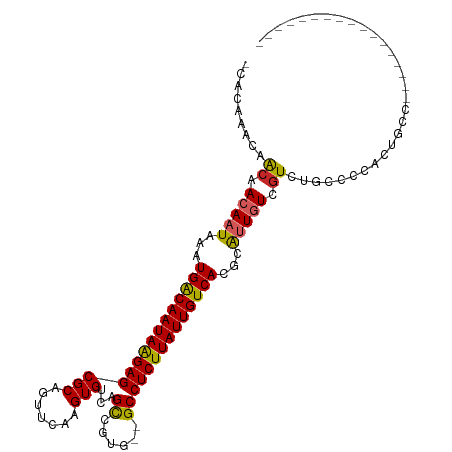
| Location | 15,268,866 – 15,268,957 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.54 |
| Shannon entropy | 0.40400 |
| G+C content | 0.49912 |
| Mean single sequence MFE | -31.99 |
| Consensus MFE | -25.96 |
| Energy contribution | -26.99 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15268866 91 - 24543557 ------------------GCAGUGGGGCAGACGACAAUGCGUGACAAUAAGAGGC--CACGGCUGACACUUGAACUGCGCUCUUAUUGUCAUUUAUUGUUGUUGUUUGUGC ------------------(((...((((((.((((((((.(((((((((((((((--.....(........)....)).))))))))))))).)))))))))))))).))) ( -33.10, z-score = -2.72, R) >droAna3.scaffold_13337 5567144 104 + 23293914 GCAGUGGCAGGGGCUUGGGC--GGGGGCAGACGACAAUGCGUGACAAUAAGAGGC--CACGACUGACACUUGAACUGCGCUCUUAUUGUCAUUUAUUGUUGUCCCUAU--- ((...(((....)))...))--(((((((....((((((.(((((((((((((((--..(((.......)))....)).))))))))))))).)))))))))))))..--- ( -39.70, z-score = -2.88, R) >droEre2.scaffold_4784 17529495 92 - 25762168 -----------------GGCAGUGAGGCAGACGACAAUGCGUGACAAUAAGAGGC--CACGGCUGACACUUGAACUGCGCUCUUAUUGUCAUUUAUUGUUGUUGUUUGUGC -----------------.(((...((((((.((((((((.(((((((((((((((--.....(........)....)).))))))))))))).)))))))))))))).))) ( -33.20, z-score = -2.73, R) >droYak2.chr3L 17780475 92 - 24197627 -----------------GGUAGUGGGGCAGACGACAAUGCGUGACAAUAAGAGGC--CACGGCUGACACUUGAACUGCGCUCUUAUUGUCAUUUAUUGUUGUUGUUUGUGC -----------------.......(.(((((((((((((.(((((((((((((((--.....(........)....)).)))))))))))))....))))))))))))).) ( -32.50, z-score = -2.70, R) >droSec1.super_0 7380903 91 - 21120651 ------------------GCAGUGGGGCAGACGACAAUGCGUGACAAUAAGAGGC--CACGGCUGACACUUGAACUGCGCUCUUAUUGUCAUUUAUUGUUGUUGUUUGUGC ------------------(((...((((((.((((((((.(((((((((((((((--.....(........)....)).))))))))))))).)))))))))))))).))) ( -33.10, z-score = -2.72, R) >droSim1.chr3L 14609223 91 - 22553184 ------------------GCAGUGGGGCAGACGACAAUGCGUGACAAUAAGAGGC--CACGGCUGACACUUGAACUGCGCUCUUAUUGUCAUUUAUUGUUGUUGUUUGCGC ------------------........(((((((((((((.(((((((((((((((--.....(........)....)).)))))))))))))....))))))))))))).. ( -34.10, z-score = -2.93, R) >droGri2.scaffold_15110 4454391 83 + 24565398 ----------------GGGC--CAGGGGGUAGGACAACACGUGACAAUAAGAGGC--CACGGCUGACACUUGAACUGCGCUCUUAUUGUCAUUUAUUGUUGUU-------- ----------------..((--(....)))..(((((((.(((((((((((((((--.....(........)....)).)))))))))))))....)))))))-------- ( -26.70, z-score = -2.18, R) >droWil1.scaffold_180955 2172800 95 + 2875958 ----------GGUAACUGAAGGUGGGGCAGACGACAACGCGUGACAAUAAGAGGCCACACGGCUGACACUUGAACUGCGCUCUUAUUGCCAUUUAUUGUUGCUUU------ ----------((((((...((((((.....(((......)))..((((((((((((....)))...(....).......)))))))))))))))...))))))..------ ( -24.50, z-score = -0.42, R) >dp4.chrXR_group6 12831228 108 + 13314419 GGACUGGUGCGGUCUGGGACUUGGGGGCAGACGACAAUGCGUGACAAUAAGAGGC--CACGGCUGACACUUGAACUGCGCUCCUAUUGUCAUUUAGUUUUGUUGCUGUUG- ((((((...))))))..........(((((.((((((.(((((((((((.(((((--.....(........)....)).))).)))))))))...)).))))))))))).- ( -31.50, z-score = -0.00, R) >droPer1.super_12 1250984 108 + 2414086 GGACUGGUGCGGUCUGGGACUUGGGGGCAGACGACAAUGCGUGACAAUAAGAGGC--CACGGCUGACACUUGAACUGCGCUCCUAUUGUCAUUUAGUUUUGUUGCUGUUG- ((((((...))))))..........(((((.((((((.(((((((((((.(((((--.....(........)....)).))).)))))))))...)).))))))))))).- ( -31.50, z-score = -0.00, R) >consensus _________________GGCAGUGGGGCAGACGACAAUGCGUGACAAUAAGAGGC__CACGGCUGACACUUGAACUGCGCUCUUAUUGUCAUUUAUUGUUGUUGUUUGUG_ .............................((((((((((.(((((((((((((((..((.(.......).))....)).))))))))))))).))))))))))........ (-25.96 = -26.99 + 1.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:28:20 2011