| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,259,033 – 15,259,140 |
| Length | 107 |
| Max. P | 0.919296 |

| Location | 15,259,033 – 15,259,140 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.39 |
| Shannon entropy | 0.30951 |
| G+C content | 0.47523 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -17.24 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.608873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 15259033 107 + 24543557 UUAGAUCUCGAGAGUA-----UCCCAUCUCUUUUAAUGGUGUACGCUUAUCCCACCGGAGUGAGCUCCCAUUUAAGCACACACCUGAGAAAGGAUCCCCCAAAUGAACCAUC .....((((((((...-----.....)))).......(((((..(((((.......((((....))))....)))))..))))).))))..((.((........)).))... ( -26.80, z-score = -2.51, R) >droEre2.scaffold_4784 17519566 104 + 25762168 UUAGAACUCGGGAACA-----UCGUAUCUAUUUUAAUG-UGUGCGCUUAUCCCACCGGUGUGAGCUCCCAUUUAGGCAGACACCUGAGA--GGAUCCCCCAAAUGAAGCAUC ......(((((((...-----.(((((.(((....)))-.)))))....))))...(((((..(((........)))..))))).))).--((.....))............ ( -24.70, z-score = -0.24, R) >droYak2.chr3L 17770109 107 + 24197627 UUAGAACUCGAGAACA-----UCCUAUCUCAUUUAAUGGCGUACGCUUAUCCCAGCGGAGUGAGCUCCCAUUUGAGCACACACCUGAGAAAGGAUCCCUCAAAUGAACCAUC .........(((...(-----((((.(((((.....(((.(((....))).)))..((.(((.((((......)))).))).))))))).)))))..)))............ ( -29.10, z-score = -2.91, R) >droSec1.super_0 7371136 112 + 21120651 CUAGAGCCUGGGAGCAUCUCUUCCCAUCUCUUUUAAUGGUGUACGCUUAUCCCACCGGAGUGAGCUCCCAUUUGAGCACACACCUGAGAAAGGAUCCUCCAAAUGAACCAUC ...(((...(((((......)))))(((.(((((...(((((..(((((.......((((....))))....)))))..)))))...)))))))).)))............. ( -29.30, z-score = -0.88, R) >droSim1.chr3L 14599498 102 + 22553184 -------CUAGAGGAG---CAUACCAUCUCUUUUAAUGGUGUACGCUUAUCCCAACGGAGUGAGCUCCCAUUUGAGCACACACCUGAGAAAAGAUCCUCCAAAUGAACCAUC -------...((((((---.(((((((........))))))).).(((.((.((..((.(((.((((......)))).))).)))).)).))).)))))............. ( -27.30, z-score = -2.61, R) >consensus UUAGAACUCGGGAGCA_____UCCCAUCUCUUUUAAUGGUGUACGCUUAUCCCACCGGAGUGAGCUCCCAUUUGAGCACACACCUGAGAAAGGAUCCCCCAAAUGAACCAUC .........................((((.((((...(((((..(((((.......((((....))))....)))))..)))))..)))).))))................. (-17.24 = -17.68 + 0.44)


| Location | 15,259,033 – 15,259,140 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.39 |
| Shannon entropy | 0.30951 |
| G+C content | 0.47523 |
| Mean single sequence MFE | -36.41 |
| Consensus MFE | -25.38 |
| Energy contribution | -25.42 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
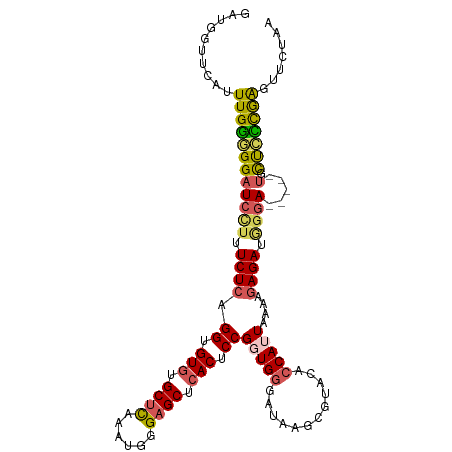
>dm3.chr3L 15259033 107 - 24543557 GAUGGUUCAUUUGGGGGAUCCUUUCUCAGGUGUGUGCUUAAAUGGGAGCUCACUCCGGUGGGAUAAGCGUACACCAUUAAAAGAGAUGGGA-----UACUCUCGAGAUCUAA ..(((.((..((((((((((((.((((.((((((((((((.((.((((....)))).))....)))))))))))).......)))).))))-----).))))))))).))). ( -39.80, z-score = -3.21, R) >droEre2.scaffold_4784 17519566 104 - 25762168 GAUGCUUCAUUUGGGGGAUCC--UCUCAGGUGUCUGCCUAAAUGGGAGCUCACACCGGUGGGAUAAGCGCACA-CAUUAAAAUAGAUACGA-----UGUUCCCGAGUUCUAA .......((((((((((((((--.....)).)))).))))))))(((((((((....))((((....((....-..............)).-----...))))))))))).. ( -27.97, z-score = -0.02, R) >droYak2.chr3L 17770109 107 - 24197627 GAUGGUUCAUUUGAGGGAUCCUUUCUCAGGUGUGUGCUCAAAUGGGAGCUCACUCCGCUGGGAUAAGCGUACGCCAUUAAAUGAGAUAGGA-----UGUUCUCGAGUUCUAA ..(((...((((((((((((((.(((((((.(((.((((......)))).))).))(((......))).............))))).))))-----).))))))))).))). ( -39.60, z-score = -3.09, R) >droSec1.super_0 7371136 112 - 21120651 GAUGGUUCAUUUGGAGGAUCCUUUCUCAGGUGUGUGCUCAAAUGGGAGCUCACUCCGGUGGGAUAAGCGUACACCAUUAAAAGAGAUGGGAAGAGAUGCUCCCAGGCUCUAG ..(((..(....((((.(((.(((((((((.(((.((((......)))).))).))((((...........))))...........))))))).))).))))...)..))). ( -38.00, z-score = -1.22, R) >droSim1.chr3L 14599498 102 - 22553184 GAUGGUUCAUUUGGAGGAUCUUUUCUCAGGUGUGUGCUCAAAUGGGAGCUCACUCCGUUGGGAUAAGCGUACACCAUUAAAAGAGAUGGUAUG---CUCCUCUAG------- ...........(((((((.(((.(((((((.(((.((((......)))).))).))..))))).))).(((.((((((......)))))).))---)))))))).------- ( -36.70, z-score = -3.06, R) >consensus GAUGGUUCAUUUGGGGGAUCCUUUCUCAGGUGUGUGCUCAAAUGGGAGCUCACUCCGGUGGGAUAAGCGUACACCAUUAAAAGAGAUGGGA_____UGCUCCCGAGUUCUAA .........((((((((.((((.((((.((.(((.((((......)))).))).))(((((............)))))....)))).)))).......))))))))...... (-25.38 = -25.42 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:28:18 2011