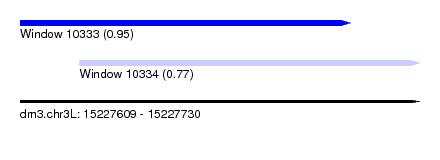
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 15,227,609 – 15,227,730 |
| Length | 121 |
| Max. P | 0.952501 |

| Location | 15,227,609 – 15,227,709 |
|---|---|
| Length | 100 |
| Sequences | 10 |
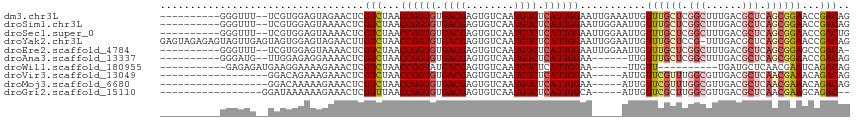
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.00 |
| Shannon entropy | 0.49751 |
| G+C content | 0.44809 |
| Mean single sequence MFE | -27.39 |
| Consensus MFE | -17.96 |
| Energy contribution | -18.01 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
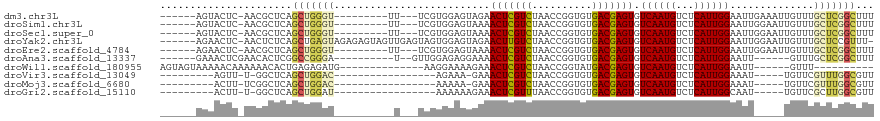
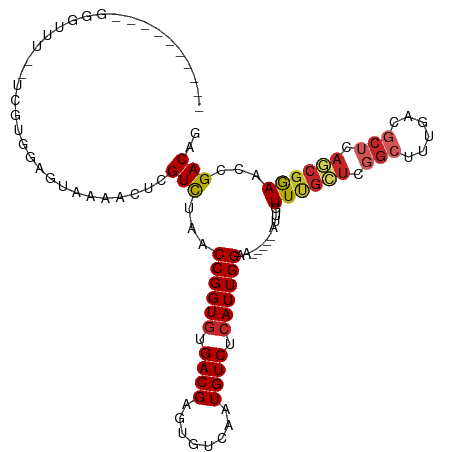
>dm3.chr3L 15227609 100 + 24543557 ------AGUACUC-AACGCUCAGCUGGGU---------UU---UCGUGGAGUAGAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAAUUGAAAUUGUUUGCUCGGCUUU ------.(.((((-(.(.....).)))))---------..---.)((.((((((((((((((..........))))))).(((((..(.....)..))))).....))))))).))... ( -27.10, z-score = -0.61, R) >droSim1.chr3L 14565731 100 + 22553184 ------AGUACUC-AACGCUCAGCUGGGU---------UU---UCGUGGAGUAAAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAAUUGGAAUUGUUUGCUCGGCUUU ------.(.((((-(.(.....).)))))---------..---.)((.(((((((.....(((((((((((.((((........)))).))))))..)))))....))))))).))... ( -28.30, z-score = -1.21, R) >droSec1.super_0 7339771 100 + 21120651 ------AGUACUC-AACGCUCAGCUGGGU---------UU---UCGUGGAGUAAAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAAUUGGAAUUGUUUGCUCGGCUUU ------.(.((((-(.(.....).)))))---------..---.)((.(((((((.....(((((((((((.((((........)))).))))))..)))))....))))))).))... ( -28.30, z-score = -1.21, R) >droYak2.chr3L 17737518 111 + 24197627 ------AGAACUC-AACUCUCAGCUGAGUAGAGAGUAGUUGAGUAGUGGAGUAGAACUUGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAAUUGGAAUUGUUUGCUCCGUUU- ------...((((-(((((((...........))).))))))))...((((((((.....(((((((((((.((((........)))).))))))..)))))....))))))))....- ( -37.60, z-score = -3.03, R) >droEre2.scaffold_4784 17488552 100 + 25762168 ------AGAACUC-AACGCUCAGCUGGGU---------UU---UCGUGGAGUAAAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAAUUGGAAUUGUUUGCUCGGCUUU ------(((((((-(.(.....).)))))---------))---).((.(((((((.....(((((((((((.((((........)))).))))))..)))))....))))))).))... ( -30.80, z-score = -1.89, R) >droAna3.scaffold_13337 5528199 95 - 23293914 ------GAAACUCGAACACUCGGCCGGGA----------U--GUUGGAGAGGAAAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAAUU------GUUUGCUCGGCUUU ------......(((....)))((((((.----------.--.((((((((.....))).)))))((((((.((((........)))).))))))....------.....))))))... ( -28.70, z-score = -1.01, R) >droWil1.scaffold_180955 2126385 89 - 2875958 AGUAGUAAAAACAAAAAACACUGAGAGAUG--------------AAGGAAAAGAAACUCGUCUAACCGGUAUGACGAGUGUCAAUGUCUCAUUGGAAUU------GUUU---------- ........((((((....((.(((((.((.--------------...((......(((((((..........))))))).)).)).))))).))...))------))))---------- ( -21.30, z-score = -2.28, R) >droVir3.scaffold_13049 20015816 85 + 25233164 ---------AGUU-U-GGCUCAGCUGGAC-----------------AGAAA-GAAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAAAU-----UGUUCGUUUGGCGUU ---------....-.-.(((.(((.((((-----------------((...-...(((((((..........))))))).((((((...))))))...)-----)))))))).)))... ( -23.10, z-score = -0.74, R) >droMoj3.scaffold_6680 7720140 86 + 24764193 ---------ACUU-UCGGCUCAGCUGGAC-----------------AAAAA-GAAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAAAU-----UGUUCGUUUGGCGUU ---------....-...(((.(((.((((-----------------((...-...(((((((..........))))))).((((((...))))))...)-----)))))))).)))... ( -23.40, z-score = -1.09, R) >droGri2.scaffold_15110 4414348 86 - 24565398 ---------ACUU-U-GGCUCAGCUGGAU-----------------AAAAAAGAAACUCGUUUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGCAAU-----UGUUCGCUUGGCGUU ---------....-.-.(((.(((.((((-----------------((.......(((((((..........)))))))(((((((...)))))))..)-----)))))))).)))... ( -25.30, z-score = -1.93, R) >consensus ______AGAACUC_AACGCUCAGCUGGGU__________U___UCGUGGAGUAAAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAAUU_____UGUUUGCUCGGCUUU ......................(((((((..........................(((((((..........))))))).((((((...))))))..............)))))))... (-17.96 = -18.01 + 0.05)
| Location | 15,227,627 – 15,227,730 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.58 |
| Shannon entropy | 0.43068 |
| G+C content | 0.47805 |
| Mean single sequence MFE | -30.01 |
| Consensus MFE | -19.70 |
| Energy contribution | -20.05 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.766161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
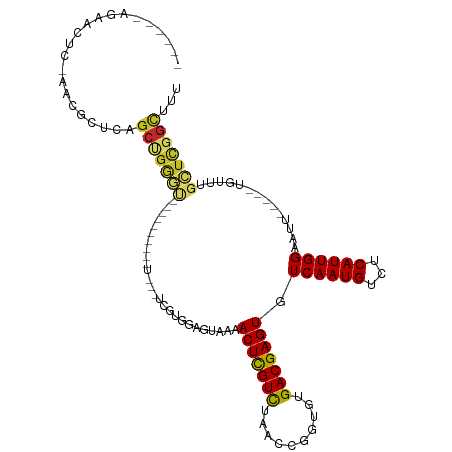
>dm3.chr3L 15227627 103 + 24543557 ----------GGGUUU--UCGUGGAGUAGAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAAUUGAAAUUGUUUGCUCGGCUUUGACGCUCAGCGGAACCGACAG ----------.(((((--.(((.((((...(((((((..........)))))))(((((.((((((.......)))....(....)..))).))))))))).))))))))..... ( -30.50, z-score = -0.55, R) >droSim1.chr3L 14565749 103 + 22553184 ----------GGGUUU--UCGUGGAGUAAAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAAUUGGAAUUGUUUGCUCGGCUUUGACGCUCAGCGGAACCGACAG ----------.(((((--..((.(((((((.....(((((((((((.((((........)))).))))))..)))))....))))))).)).....(((...))))))))..... ( -31.70, z-score = -1.03, R) >droSec1.super_0 7339789 103 + 21120651 ----------GGGUUU--UCGUGGAGUAAAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAAUUGGAAUUGUUUGCUCGGCUUUGACGCUCAGCGGAACCGACUG ----------.(((((--..((.(((((((.....(((((((((((.((((........)))).))))))..)))))....))))))).)).....(((...))))))))..... ( -31.70, z-score = -1.18, R) >droYak2.chr3L 17737536 114 + 24197627 GAGUAGAGAGUAGUUGAGUAGUGGAGUAGAACUUGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAAUUGGAAUUGUUUGCUCCG-UUUGACGCUCAGCGGAACCGACAG .......((((.((..((...(((((((((.....(((((((((((.((((........)))).))))))..)))))....)))))))))-))..)))))).(((....)).).. ( -34.00, z-score = -1.37, R) >droEre2.scaffold_4784 17488570 102 + 25762168 ----------GGGUUU--UCGUGGAGUAAAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAAUUGGAAUUGUUUGCUCGGCUUUGACGCUCAGCGGAGCCGACA- ----------..(((.--.....(((((((.....(((((((((((.((((........)))).))))))..)))))....)))))))(((((((.(.....))))))))))).- ( -32.40, z-score = -1.01, R) >droAna3.scaffold_13337 5528218 97 - 23293914 ----------GGGAUG--UUGGAGAGGAAAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAA------UUGUUUGCUCGGCUUUGACGCUCAGCGGAACCGACAG ----------....((--(((((((.((....)).)))..((((((.((((........)))).))))))..------...((((((.(((......))).)))))).)))))). ( -30.70, z-score = -0.89, R) >droWil1.scaffold_180955 2126407 88 - 2875958 -----------GAGAGAUGAAGGAAAAGAAACUCGUCUAACCGGUAUGACGAGUGUCAAUGUCUCAUUGGAA------UUGUU----------UGAUGCUCAACGAAUCAGACAG -----------((((.((....((......(((((((..........))))))).)).)).)))).......------.((((----------((((.........)))))))). ( -23.30, z-score = -1.45, R) >droVir3.scaffold_13049 20015830 92 + 25233164 ------------------GGACAGAAAGAAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAA-----AUUGUUCGUUUGGCGUUGACGCUCAACGAAACAGACAG ------------------((((((......(((((((..........))))))).((((((...))))))..-----.))))))(((((.(((((.....)))))...))))).. ( -29.40, z-score = -1.88, R) >droMoj3.scaffold_6680 7720155 92 + 24764193 ------------------GGACAAAAAGAAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAA-----AUUGUUCGUUUGGCGUUGACGCUCAACGAAACAGACAG ------------------((((((......(((((((..........))))))).((((((...))))))..-----.))))))(((((.(((((.....)))))...))))).. ( -29.70, z-score = -2.31, R) >droGri2.scaffold_15110 4414362 91 - 24565398 -----------------GGAUAAAAAAGAAACUCGUUUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGCA-----AUUGUUCGCUUGGCGUUGACGCUCAACGAAGCAGAC-- -----------------((((((.......(((((((..........)))))))(((((((...))))))).-----.))))))(((...(((((.....))))).)))....-- ( -26.70, z-score = -1.51, R) >consensus __________GGGUUU__UCGUGGAGUAAAACUCGUCUAACCGGUGUGACGAGUGUCAAUGUCUCAUUGGAA_____AUUGUUUGCUCGGCUUUGACGCUCAGCGGAACCGACAG ..................................(((...((((((.((((........)))).))))))...........((((((.(((......))).))))))...))).. (-19.70 = -20.05 + 0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:28:11 2011